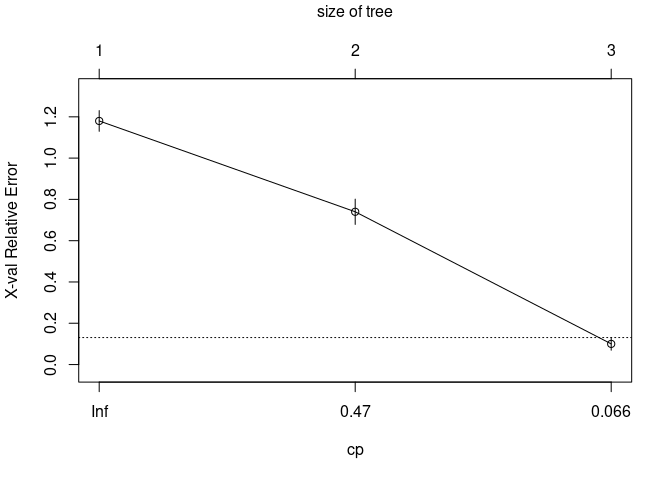
They create different objects and use surrogate variables differently.
library(rpart)
iris_tree <- rpart(formula = Species ~ Sepal.Length + Sepal.Width + Petal.Length + Petal.Width, method="class", data=iris)
plot(iris_tree)
str(iris_tree)
#> List of 14
#> $ frame :'data.frame': 5 obs. of 9 variables:
#> ..$ var : chr [1:5] "Petal.Length" "<leaf>" "Petal.Width" "<leaf>" ...
#> ..$ n : int [1:5] 150 50 100 54 46
#> ..$ wt : num [1:5] 150 50 100 54 46
#> ..$ dev : num [1:5] 100 0 50 5 1
#> ..$ yval : num [1:5] 1 1 2 2 3
#> ..$ complexity: num [1:5] 0.5 0.01 0.44 0 0.01
#> ..$ ncompete : int [1:5] 3 0 3 0 0
#> ..$ nsurrogate: int [1:5] 3 0 3 0 0
#> ..$ yval2 : num [1:5, 1:8] 1 1 2 2 3 50 50 0 0 0 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:8] "" "" "" "" ...
#> $ where : Named int [1:150] 2 2 2 2 2 2 2 2 2 2 ...
#> ..- attr(*, "names")= chr [1:150] "1" "2" "3" "4" ...
#> $ call : language rpart(formula = Species ~ Sepal.Length + Sepal.Width + Petal.Length + Petal.Width, data = iris, method = "class")
#> $ terms :Classes 'terms', 'formula' language Species ~ Sepal.Length + Sepal.Width + Petal.Length + Petal.Width
#> .. ..- attr(*, "variables")= language list(Species, Sepal.Length, Sepal.Width, Petal.Length, Petal.Width)
#> .. ..- attr(*, "factors")= int [1:5, 1:4] 0 1 0 0 0 0 0 1 0 0 ...
#> .. .. ..- attr(*, "dimnames")=List of 2
#> .. .. .. ..$ : chr [1:5] "Species" "Sepal.Length" "Sepal.Width" "Petal.Length" ...
#> .. .. .. ..$ : chr [1:4] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
#> .. ..- attr(*, "term.labels")= chr [1:4] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
#> .. ..- attr(*, "order")= int [1:4] 1 1 1 1
#> .. ..- attr(*, "intercept")= int 1
#> .. ..- attr(*, "response")= int 1
#> .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
#> .. ..- attr(*, "predvars")= language list(Species, Sepal.Length, Sepal.Width, Petal.Length, Petal.Width)
#> .. ..- attr(*, "dataClasses")= Named chr [1:5] "factor" "numeric" "numeric" "numeric" ...
#> .. .. ..- attr(*, "names")= chr [1:5] "Species" "Sepal.Length" "Sepal.Width" "Petal.Length" ...
#> $ cptable : num [1:3, 1:5] 0.5 0.44 0.01 0 1 2 1 0.5 0.06 1.16 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : chr [1:3] "1" "2" "3"
#> .. ..$ : chr [1:5] "CP" "nsplit" "rel error" "xerror" ...
#> $ method : chr "class"
#> $ parms :List of 3
#> ..$ prior: num [1:3(1d)] 0.333 0.333 0.333
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:3] "1" "2" "3"
#> ..$ loss : num [1:3, 1:3] 0 1 1 1 0 1 1 1 0
#> ..$ split: num 1
#> $ control :List of 9
#> ..$ minsplit : int 20
#> ..$ minbucket : num 7
#> ..$ cp : num 0.01
#> ..$ maxcompete : int 4
#> ..$ maxsurrogate : int 5
#> ..$ usesurrogate : int 2
#> ..$ surrogatestyle: int 0
#> ..$ maxdepth : int 30
#> ..$ xval : int 10
#> $ functions :List of 3
#> ..$ summary:function (yval, dev, wt, ylevel, digits)
#> ..$ print :function (yval, ylevel, digits)
#> ..$ text :function (yval, dev, wt, ylevel, digits, n, use.n)
#> $ numresp : int 5
#> $ splits : num [1:14, 1:5] 150 150 150 150 0 0 0 100 100 100 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : chr [1:14] "Petal.Length" "Petal.Width" "Sepal.Length" "Sepal.Width" ...
#> .. ..$ : chr [1:5] "count" "ncat" "improve" "index" ...
#> $ variable.importance: Named num [1:4] 89 81.3 54.1 36
#> ..- attr(*, "names")= chr [1:4] "Petal.Width" "Petal.Length" "Sepal.Length" "Sepal.Width"
#> $ y : int [1:150] 1 1 1 1 1 1 1 1 1 1 ...
#> $ ordered : Named logi [1:4] FALSE FALSE FALSE FALSE
#> ..- attr(*, "names")= chr [1:4] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
#> - attr(*, "xlevels")= Named list()
#> - attr(*, "ylevels")= chr [1:3] "setosa" "versicolor" "virginica"
#> - attr(*, "class")= chr "rpart"
library(DAAG)
#> Loading required package: lattice
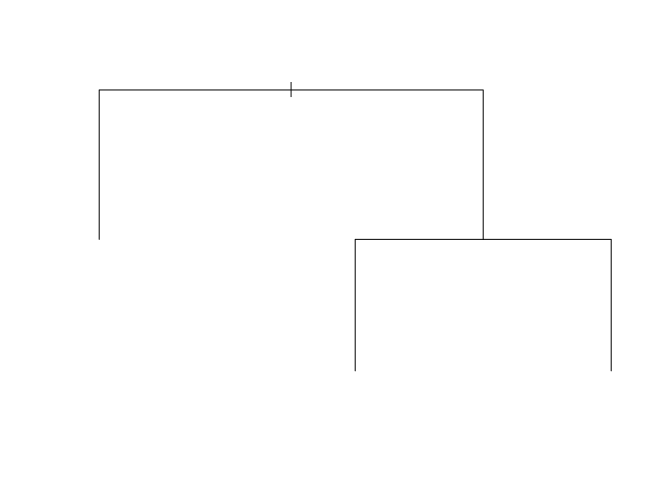
library(tree)
iris_tr <- tree(Species ~., iris )
plot(iris_tr)
str(iris_tr)
#> List of 6
#> $ frame :'data.frame': 11 obs. of 6 variables:
#> ..$ var : Factor w/ 5 levels "<leaf>","Sepal.Length",..: 4 1 5 4 2 1 1 1 4 1 ...
#> ..$ n : num [1:11] 150 50 100 54 48 5 43 6 46 6 ...
#> ..$ dev : num [1:11] 329.58 0 138.63 33.32 9.72 ...
#> ..$ yval : Factor w/ 3 levels "setosa","versicolor",..: 1 1 2 2 2 2 2 3 3 3 ...
#> ..$ splits: chr [1:11, 1:2] "<2.45" "" "<1.75" "<4.95" ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:2] "cutleft" "cutright"
#> ..$ yprob : num [1:11, 1:3] 0.333 1 0 0 0 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:3] "setosa" "versicolor" "virginica"
#> $ where : Named int [1:150] 2 2 2 2 2 2 2 2 2 2 ...
#> ..- attr(*, "names")= chr [1:150] "1" "2" "3" "4" ...
#> $ terms :Classes 'terms', 'formula' language Species ~ Sepal.Length + Sepal.Width + Petal.Length + Petal.Width
#> .. ..- attr(*, "variables")= language list(Species, Sepal.Length, Sepal.Width, Petal.Length, Petal.Width)
#> .. ..- attr(*, "factors")= int [1:5, 1:4] 0 1 0 0 0 0 0 1 0 0 ...
#> .. .. ..- attr(*, "dimnames")=List of 2
#> .. .. .. ..$ : chr [1:5] "Species" "Sepal.Length" "Sepal.Width" "Petal.Length" ...
#> .. .. .. ..$ : chr [1:4] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
#> .. ..- attr(*, "term.labels")= chr [1:4] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
#> .. ..- attr(*, "order")= int [1:4] 1 1 1 1
#> .. ..- attr(*, "intercept")= int 1
#> .. ..- attr(*, "response")= int 1
#> .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
#> .. ..- attr(*, "predvars")= language list(Species, Sepal.Length, Sepal.Width, Petal.Length, Petal.Width)
#> .. ..- attr(*, "dataClasses")= Named chr [1:5] "factor" "numeric" "numeric" "numeric" ...
#> .. .. ..- attr(*, "names")= chr [1:5] "Species" "Sepal.Length" "Sepal.Width" "Petal.Length" ...
#> $ call : language tree(formula = Species ~ ., data = iris)
#> $ y : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
#> ..- attr(*, "names")= chr [1:150] "1" "2" "3" "4" ...
#> $ weights: num [1:150] 1 1 1 1 1 1 1 1 1 1 ...
#> - attr(*, "class")= chr "tree"
#> - attr(*, "xlevels")=List of 4
#> ..$ Sepal.Length: NULL
#> ..$ Sepal.Width : NULL
#> ..$ Petal.Length: NULL
#> ..$ Petal.Width : NULL
#> - attr(*, "ylevels")= chr [1:3] "setosa" "versicolor" "virginica"
# from help(rpart)
# This differs from the tree function in S mainly in its handling of surrogate variables. In most details it follows Breiman et. al (1984) quite closely. R package tree provides a re-implementation of tree.
# from the long intro vignette for rpart
# Once a splitting variable and a split point for it have been decided, what is to be done with observations missing that variable? One approach is to estimate the missing datum using the other independent variables; rpart uses a variation of this to define surrogate variables.
Created on 2022-12-08 by the reprex package (v2.0.1)
![]()