Hi All,
Here is my code:
irrigacje <- structure(list(irrigationes = structure(c(1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L), levels = c("Control", "Irrigated 10\nmm",
"Irrigated 20 mm"), class = "factor", contrasts = structure(c(0,
1, 0, 0, 0, 1), dim = 3:2, dimnames = list(c("Control", "Irrigated 10\nmm",
"Irrigated 20 mm"), c("Irrigated 10\nmm", "Irrigated 20 mm")))),
biomass = 1:30), row.names = c(NA, -30L), class = "data.frame")
# Calculate group statistics
group_stats <- aggregate(biomass ~ irrigationes, data = irrigacje,
function(x) c(mean = mean(x), min = min(x), max = max(x)))
group_stats in console:
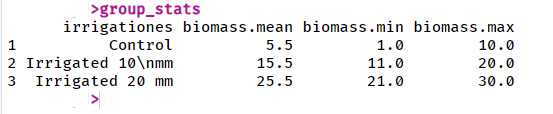
group_stats in View():
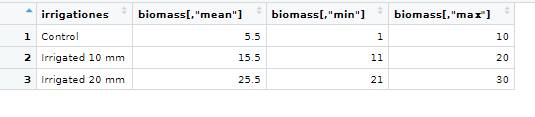
colnames(group_stats)
[1] "irrigationes" "biomass"
typeof(group_stats)
[1] "list"
class(group_stats)
[1] "data.frame"
What is going on here ? How to convert it to real dataframe that I could have proper colnames in it and export it to excel properly ?
Is it possible to edit it in RStudio ? Fix() or edit() don't work with it somehow.