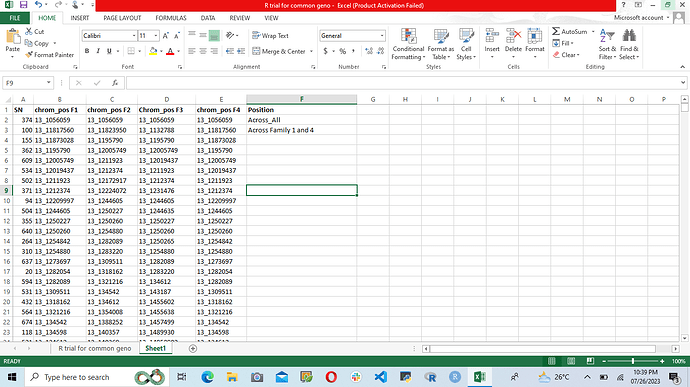
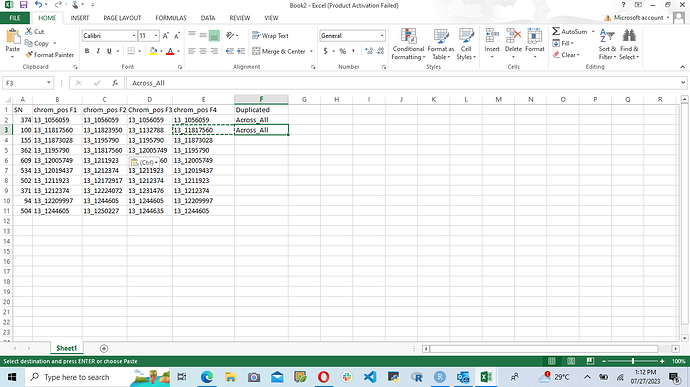
Good Evening Dear All, I have a genotyping dataset in which I want to find all the chromosomes number that can be found across the four column and I will specify that that can be found across the four column, but if that can be found in only two columns I will specify that that can be found in only two columns ...., I know that this can be achieved with across along side with case_when but I have tried it but my code is still giving me error. Here is the sampled dataset below.
library(tidyverse)
dataset
dat_geno <-
structure(
list(
SN...1 = c(
374,
100,
155,
362,
609,
534,
502,
371,
94,
504,
355,
640,
264,
310,
637,
20,
594,
531,
432,
564
),
`chrom_pos F1` = c(
"13_1056059",
"13_11817560",
"13_11873028",
"13_1195790",
"13_12005749",
"13_12019437",
"13_1211923",
"13_1212374",
"13_12209997",
"13_1244605",
"13_1250227",
"13_1250260",
"13_1254842",
"13_1254880",
"13_1273697",
"13_1282054",
"13_1282089",
"13_1309511",
"13_1318162",
"13_1321216"
),
`chrom_pos F2` = c(
"13_1056059",
"13_11823950",
"13_1195790",
"13_12005749",
"13_1211923",
"13_1212374",
"13_12172917",
"13_12224072",
"13_1244605",
"13_1250227",
"13_1250260",
"13_1254880",
"13_1282089",
"13_1283220",
"13_1309511",
"13_1318162",
"13_1321216",
"13_134542",
"13_134612",
"13_1354008"
),
`Chrom_pos F3` = c(
"13_1056059",
"13_1132788",
"13_1195790",
"13_12005749",
"13_12019437",
"13_1211923",
"13_1212374",
"13_1231476",
"13_1244605",
"13_1244635",
"13_1250227",
"13_1250260",
"13_1250265",
"13_1254880",
"13_1282089",
"13_1283220",
"13_134612",
"13_143187",
"13_1455602",
"13_1455638"
),
`chrom_pos F4` = c(
"13_1056059",
"13_11817560",
"13_11873028",
"13_1195790",
"13_12005749",
"13_12019437",
"13_1211923", "13_1212374", "13_12209997", "13_1244605", "13_1250227",
"13_1250260", "13_1254842", "13_1254880", "13_1273697", "13_1282054",
"13_1282089", "13_1309511", "13_1318162", "13_1321216")), row.names = c(NA,
-20L), class = c("tbl_df", "tbl", "data.frame"))