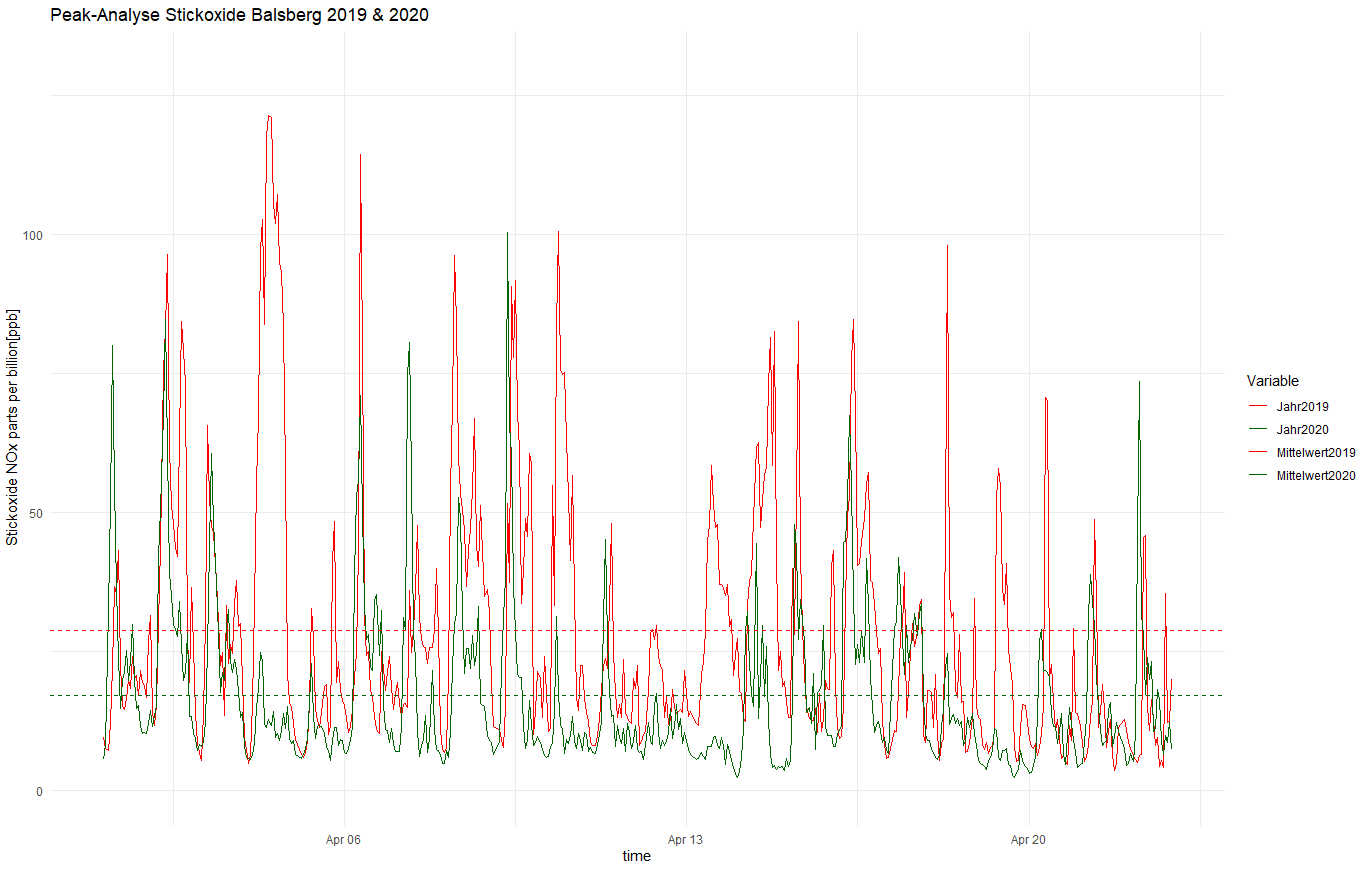
Hi All
I work with ggplot and I have a problem with the legend.
The dashed lines in the plot are not dashed in the legend, even though I defined their style with the command: scale_linetype_manual
Suspicious is also, I do not receive a warning or error in the console. However, it is not executed.
Here is my code, followed by the plot:
ggplot(nox, aes(x=Zeit))+
geom_line(aes(y=Jahr2019,color="Jahr2019"))+
geom_line(aes(y=Jahr2020,color="Jahr2020"))+
geom_hline(aes(color="Mittelwert2019",yintercept=28.72),linetype="dashed",size=0.5)+
geom_hline(aes(color="Mittelwert2020",yintercept=17.01),linetype="dashed",size=0.5)+
ylim(0,130)+
ggtitle("Peak-Analyse Stickoxide Balsberg 2019 & 2020")+
labs(x="time",
y=" Stickoxide NOx parts per billion[ppb]")+
scale_color_manual(name="Variable",
breaks = c("Jahr2019", "Jahr2020", "Mittelwert2019", "Mittelwert2020"),
values = c("Jahr2019" = "red", "Jahr2020" = "darkgreen", "Mittelwert2019"="Red","Mittelwert2020"="Darkgreen"))+
scale_linetype_manual(breaks = c("Jahr2019", "Jahr2020", "Mittelwert2019", "Mittelwert2020"),
values = c("Jahr2019" = "solid", "Jahr2020" = "solid", "Mittelwert2019"="dashed","Mittelwert2020"="dashed"))+
theme_minimal()
Thank you very much for your help. The problem seems easy but somehow I struggle with it