I have a data frame that contains ranges (defined by their start and end), and a second data frame that contains cut points. There can be 0, 1, or several cut points within each range. Whenever a cut point falls within a range, I want to separate that range in two rows, using the cut point as end and start of the new rows.
Illustration with fake data:
suppressPackageStartupMessages(library(tidyverse))
source_df <- tibble(id = LETTERS[1:4],
start = as.integer(10*(1:4)),
end = as.integer(10*(1:4)+5))
cutting_df <- tibble(cutpoint = c(12L, 23L, 32L,33L))
expected_result <- tibble(id = c("A1","A2","B1","B2","C1","C2","C3","D"),
start = c( 10L, 12L, 20L, 23L, 30L, 32L, 33L, 40L),
end = c( 12L, 15L, 23L, 25L, 32L, 33L, 35L, 45L))
source_df
#> # A tibble: 4 x 3
#> id start end
#> <chr> <int> <int>
#> 1 A 10 15
#> 2 B 20 25
#> 3 C 30 35
#> 4 D 40 45
cutting_df
#> # A tibble: 4 x 1
#> cutpoint
#> <int>
#> 1 12
#> 2 23
#> 3 32
#> 4 33
expected_result
#> # A tibble: 8 x 3
#> id start end
#> <chr> <int> <int>
#> 1 A1 10 12
#> 2 A2 12 15
#> 3 B1 20 23
#> 4 B2 23 25
#> 5 C1 30 32
#> 6 C2 32 33
#> 7 C3 33 35
#> 8 D 40 45
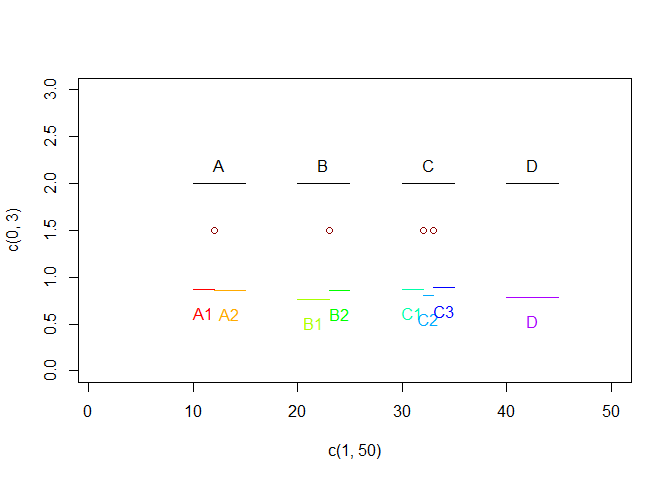
plot(c(1,50), c(0,3), col="white")
for(i in seq_len(nrow(source_df))){
lines(c(source_df[i, "start"], source_df[i, "end"]), c(2,2))
text((source_df[i, "end"]-source_df[i, "start"])/2 + source_df[i, "start"], 2.2, source_df[i, "id"])
}
for(i in seq_len(nrow(cutting_df))){
points(cutting_df$cutpoint[i], 1.5, col = "darkred")
}
for(i in seq_len(nrow(expected_result))){
jitter <- runif(1,max = .2)
lines(x = c(expected_result[i, "start"], expected_result[i, "end"]),
y = c(.75,.75) + jitter,
col = rainbow(9)[i])
text(x = (expected_result[i, "end"]-expected_result[i, "start"])/2 + expected_result[i, "start"],
y = .5+jitter,
labels = expected_result[i, "id"],
col = rainbow(9)[i])
}
The numbers are guaranteed to be all integers, my data frames have at least 10s of thousands, potentially millions of rows, so performance is a concern.
I feel like there should be a dplyr/tidyverse solution, but I can't figure it out.
Side note: actually, the real data is genomic coordinates, so I'm also exploring IRanges/GRanges solutions. But in that case too, I can't come up with an approach that doesn't require looping over the data frames several times. And since I'm doing the other analyses with the tidyverse, a dplyr solution would be more elegant.