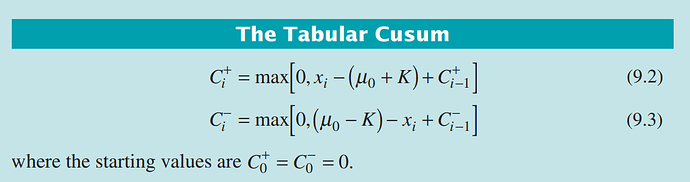
You can get the code for the function by typing cusum at the console prompt. Here is what I got.
> cusum
function (data, sizes, center, std.dev, head.start = 0, decision.interval = 5,
se.shift = 1, data.name, labels, newdata, newsizes, newlabels,
plot = TRUE, ...)
{
call <- match.call()
if (missing(data))
stop("'data' argument is not specified")
if (missing(data.name))
data.name <- deparse(substitute(data))
data <- data.matrix(data)
if (missing(sizes)) {
sizes <- apply(data, 1, function(x) sum(!is.na(x)))
}
else {
if (length(sizes) == 1)
sizes <- rep(sizes, nrow(data))
else if (length(sizes) != nrow(data))
stop("sizes length doesn't match with data")
}
if (decision.interval <= 0)
stop("decision.interval must be positive")
if (head.start < 0 || head.start >= decision.interval)
stop("head.start must be non-negative and less than decision.interval")
type <- if (any(sizes == 1))
"xbar.one"
else "xbar"
if (ncol(data) == 1 & any(sizes > 1) & missing(std.dev))
stop("sizes larger than 1 but data appears to be single samples. In this case you must provide also the std.dev")
if (missing(labels)) {
if (is.null(rownames(data)))
labels <- 1:nrow(data)
else labels <- rownames(data)
}
stats <- paste("stats.", type, sep = "")
if (!exists(stats, mode = "function"))
stop(paste("function", stats, "is not defined"))
stats <- do.call(stats, list(data, sizes))
statistics <- stats$statistics
if (missing(center))
center <- stats$center
sd <- paste("sd.", type, sep = "")
if (!exists(sd, mode = "function"))
stop(paste("function", sd, "is not defined!"))
if (missing(std.dev)) {
std.dev <- switch(type, xbar = {
if (any(sizes > 25)) "RMSDF" else "UWAVE-R"
}, NULL)
std.dev <- do.call(sd, list(data, sizes, std.dev))
}
else {
if (is.character(std.dev)) {
std.dev <- do.call(sd, list(data, sizes, std.dev))
}
else {
if (!is.numeric(std.dev))
stop("if provided the argument 'std.dev' must be a method available or a numerical value. See help(qcc).")
}
}
names(statistics) <- rownames(data) <- labels
names(dimnames(data)) <- list("Group", "Samples")
object <- list(call = call, type = "cusum", data.name = data.name,
data = data, statistics = statistics, sizes = sizes,
center = center, std.dev = std.dev)
if (!missing(newdata)) {
newdata.name <- deparse(substitute(newdata))
newdata <- data.matrix(newdata)
if (missing(newsizes)) {
newsizes <- apply(newdata, 1, function(x) sum(!is.na(x)))
}
else {
if (length(newsizes) == 1)
newsizes <- rep(newsizes, nrow(newdata))
else if (length(newsizes) != nrow(newdata))
stop("newsizes length doesn't match with newdata")
}
stats <- paste("stats.", type, sep = "")
if (!exists(stats, mode = "function"))
stop(paste("function", stats, "is not defined"))
newstats <- do.call(stats, list(newdata, newsizes))$statistics
if (missing(newlabels)) {
if (is.null(rownames(newdata))) {
start <- length(statistics)
newlabels <- seq(start + 1, start + length(newstats))
}
else {
newlabels <- rownames(newdata)
}
}
names(newstats) <- newlabels
object$newstats <- newstats
object$newdata <- newdata
object$newsizes <- newsizes
object$newdata.name <- newdata.name
statistics <- c(statistics, newstats)
sizes <- c(sizes, newsizes)
}
n <- length(statistics)
z <- (statistics - center)/(std.dev/sqrt(sizes))
ldb <- -decision.interval
udb <- decision.interval
z.f <- z - se.shift/2
cusum.pos <- rep(NA, n)
cusum.pos[1] <- max(0, head.start + z.f[1])
for (i in 2:n) cusum.pos[i] <- max(0, cusum.pos[i - 1] +
z.f[i])
z.f <- z + se.shift/2
cusum.neg <- rep(NA, n)
cusum.neg[1] <- max(0, head.start - z.f[1])
for (i in 2:n) cusum.neg[i] <- max(0, cusum.neg[i - 1] -
z.f[i])
cusum.neg <- -cusum.neg
violations <- list(lower = which(cusum.neg < ldb), upper = which(cusum.pos >
udb))
object$type <- "cusum"
object$pos <- cusum.pos
object$neg <- cusum.neg
object$head.start <- head.start
object$decision.interval <- decision.interval
object$se.shift <- se.shift
object$violations <- violations
class(object) <- "cusum.qcc"
if (plot)
plot(object, ...)
return(object)
}
<bytecode: 0x000001caa0dfec48>
<environment: namespace:qcc>