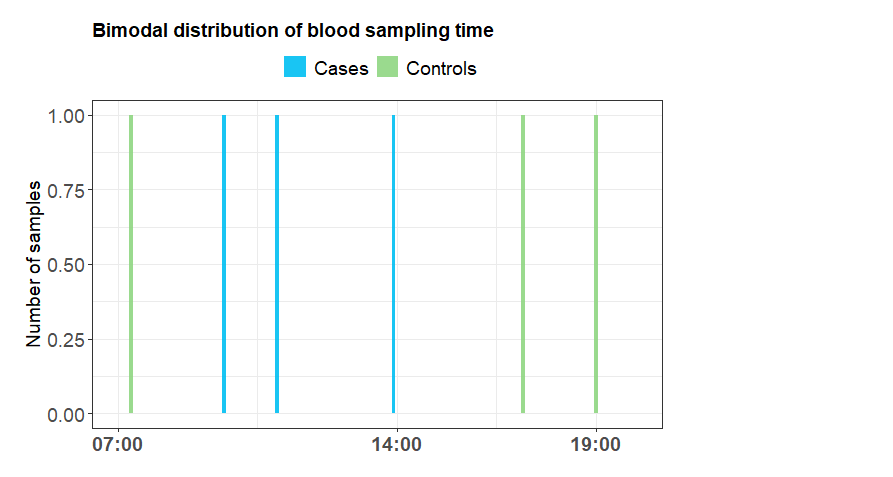
Hi all,
I have made a distribution of blood sampling times, and would like to use only three time points on the x-axis, 07:00, 14:00 and 19:00.
One problem I have is that I do not have a sample for 07:00. The first starts at 07:20. I have tried scale_x_time() without luck. Also, I would like to remove the seconds from the hms formatted time. No luck there either.
Thanks for your help.
plot <- targets2 %>%
ggplot(aes(x=Blood_draw_time, fill = factor(Case_Control))) +
geom_histogram(binwidth = 300, alpha = 0.9) +
ggtitle("Bimodal distribution of blood sampling time") +
theme_bw() +
labs(x = " ", y = "Number of samples") +
scale_fill_manual(values = c("#00bff2","#8fd682")) +
theme(
plot.title = element_text(face = "bold", size = 14),
legend.position = "top",
legend.title = element_blank(),
legend.text = element_text(size = 14),
axis.text.x = element_text(size = 14, face = "bold"),
axis.text.y = element_text(size = 14),
axis.title.y = element_text(size = 14),
plot.margin = margin(t=12, r = 14, b = 10, l = 12, unit = "pt")
)
plot
Bit of data
targets2 %>%
head() %>% tibble::tribble(
~Case_Control, ~Blood_draw_time,
"Controls", "07:20:00",
"Controls", "17:10:00",
"Controls", "19:00:00",
"Cases", "11:00:00",
"Cases", "13:57:00",
"Cases", "09:40:00"
)
dpasta()