I have a modeling workflow that uses conformal inference for prediction intervals and I'm trying to use the following workflow to deploy my models to Posit Connect:
- tune/finalize models using tidymodels workflow
- implement conformal inference using
split_int <- int_conformal_split(fitted_workflow_object, validation) - pin models using
vetiver_pin_write() - promote each pinned model to an API using
vetiver_deploy_rsconnect()
The probably docs indicate i can just use predict(split_int, new data, level= 0.90) to get prediction and intervals. My initial thought was to pass split_int into the vetiver_model, but the vetiver docs state that the model argument in vetiver_model should be a "A trained model, such as an lm() model or a tidymodels workflows::workflow()."
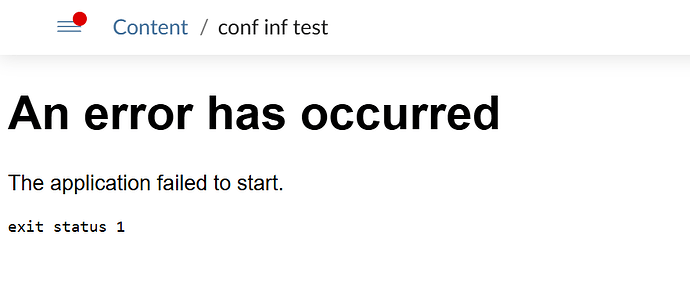
As expected when using the conformal split object as the model, it fails on the server:
and when i call the endpoint from my session i get:
Error:
! lexical error: invalid char in json text.
<!DOCTYPE html> <html lang="en-
(right here) ------^
Hide Traceback
▆
1. ├─stats::predict(...)
2. └─vetiver:::predict.vetiver_endpoint(...)
3. └─jsonlite::fromJSON(resp)
4. └─jsonlite:::parse_and_simplify(...)
5. └─jsonlite:::parseJSON(txt, bigint_as_char)
6. └─jsonlite:::parse_string(txt, bigint_as_char)
Do i need to define a custom handlers since the class of split_int is not a tuned workflow which vetiver_model expects? if so, how do i do this and where in the workflow does it fit? I check the vetiver docs on this but couldn't get very far with it.
Below is a pseudo-reprex - i wanted to mask our orgs connect endpoint:
library(workflows)
library(dplyr)
library(parsnip)
library(rsample)
library(tune)
library(modeldata)
library(probably)
library(vetiver)
library(pins)
set.seed(2)
sim_train <- sim_regression(500)
sim_cal <- sim_regression(200)
sim_new <- sim_regression(5) |> dplyr::select(-outcome)
mlp_spec <-
mlp(hidden_units = 5, penalty = 0.01) |>
set_mode("regression")
mlp_wflow <-
workflow() |>
add_model(mlp_spec) |>
add_formula(outcome ~ .)
mlp_fit <- fit(mlp_wflow, data = sim_train)
mlp_int <- probably::int_conformal_split(mlp_fit, sim_cal)
mlp_int
predict(mlp_int, sim_new, level = 0.90)
# use the int_conformal_split as the model object
v <- vetiver_model(model = mlp_int, "mlp_fit_TEST_DELETE", description = "TEST", save_prototype = FALSE)
v
board <- board_connect(auth = "envvar")
board %>% vetiver_pin_write(v)
vetiver_deploy_rsconnect(
board = board,
name = "mlp_fit_TEST_DELETE",
appTitle = "conf inf test",
predict_args = list(debug = TRUE)
)
apiKey <- Sys.getenv("CONNECT_API_KEY")
#errors out with above error
preds <-
predict(
object = endpoint, #real endpoint masked, replace with real one for reprex to work
type = "prob",
level = 0.90,
new_data = sim_new,
httr::add_headers(Authorization = paste("Key", apiKey))
)