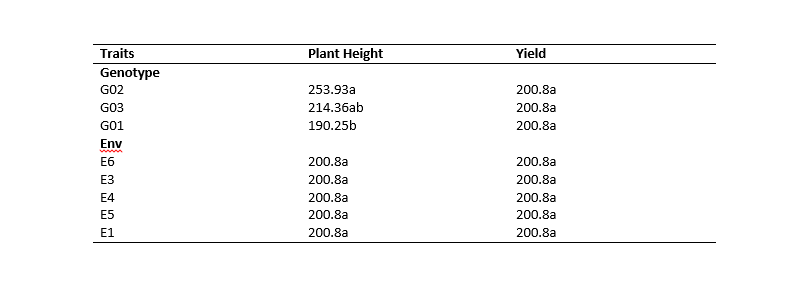
Good Morning Dear all, Please I am working on creating multiple tables for my thesis and I want to use the gt package so that. Below I attached a fake dataset and a screenshot of expected output. I will be very grateful if someone can help me out. Thanks
# Load Packages
library(gt)
library(tidyverse)
# Fake dataset
Growth <-
structure(
list(
env = structure(
c(
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
5L,
5L,
5L,
5L,
5L,
5L,
5L,
5L,
5L,
5L,
5L,
5L,
6L,
6L,
6L,
6L,
6L,
6L,
6L,
6L,
6L,
6L,
6L,
6L
),
levels = c("E1", "E2", "E3", "E4", "E5", "E6"),
class = "factor"
),
rep = structure(
c(
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
4L,
4L,
4L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
4L,
4L,
4L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
4L,
4L,
4L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
4L,
4L,
4L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
4L,
4L,
4L,
1L,
1L,
1L,
2L,
2L,
2L,
3L,
3L,
3L,
4L,
4L,
4L
),
levels = c("R1", "R2", "R3", "R4"),
class = "factor"
),
gen = structure(
c(
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L,
1L,
2L,
3L
),
levels = c(
"G01",
"G02",
"G03",
"G04",
"G05",
"G06",
"G07",
"G08",
"G09",
"G10",
"G11",
"G12",
"G13",
"G14",
"G15",
"G16",
"G17",
"G18"
),
class = "factor"
),
yield = c(
139.82,
182.52,
171.71,
141.26,
182.52,
160.97,
126.72,
182.52,
138.15,
114.52,
182.52,
166.4,
401.71,
296.22,
180.31,
210.38,
375.53,
51.14,
166.95,
337.11,
385.3,
192.17,
314.83,
231.17,
449.4,
457.84,
545.45,
440.62,
515.34,
962.56,
663.81,
625.93,
932.53,
773.59,
462.25,
779.18,
458.43,
862.26,
400.05,
192.78,
591.57,
242.55,
338.52,
604.8,
204.12,
186.69,
500.43,
242.97,
195.35,
168.85,
195.12,
195.1,
158.85,
200,
197.93,
147.01,
211.63,
199.5,
157.01,
278.85,
984.38,
984.38,
754.69,
689.06,
1220.62,
885.69,
836.72,
1594.68,
951.56,
836.68,
1082.81,
1017.19
),
`Plant Height` = c(
69.91,
91.26,
85.855,
70.63,
91.26,
80.485,
63.36,
91.26,
69.075,
57.26,
91.26,
83.2,
200.855,
148.11,
90.155,
105.19,
187.765,
25.57,
83.475,
168.555,
192.65,
96.085,
157.415,
115.585,
224.7,
228.92,
272.725,
220.31,
257.67,
481.28,
331.905,
312.965,
466.265,
386.795,
231.125,
389.59,
229.215,
431.13,
200.025,
96.39,
295.785,
121.275,
169.26,
302.4,
102.06,
93.345,
250.215,
121.485,
97.675,
84.425,
97.56,
97.55,
79.425,
100,
98.965,
73.505,
105.815,
99.75,
78.505,
139.425,
492.19,
492.19,
377.345,
344.53,
610.31,
442.845,
418.36,
797.34,
475.78,
418.34,
541.405,
508.595
)
),
class = "data.frame",
row.names = c(NA,
-72L)
)
# Sreenshot of Expected Output
Thank you very much!!!