So I'm really new to R and am trying to create some figures in RStudio but I am having a hard time getting my plots to work. I've included my code and the output below. Basically, I want to create a group bar chart where I have the information grouped by the treatment group, but then each of the "_avg" columns is one variable that I want plotted as a y. I'm not sure how to set the y and fill to reflect this. Any input is helpful.
Code:
# RSTUDIO PACKAGE INSTALLATION ################################################
library(janitor)
library(tidyverse)
library(ggplot2)
library(readr)
library(dplyr)
# IMPORT THE DATA #############################################################
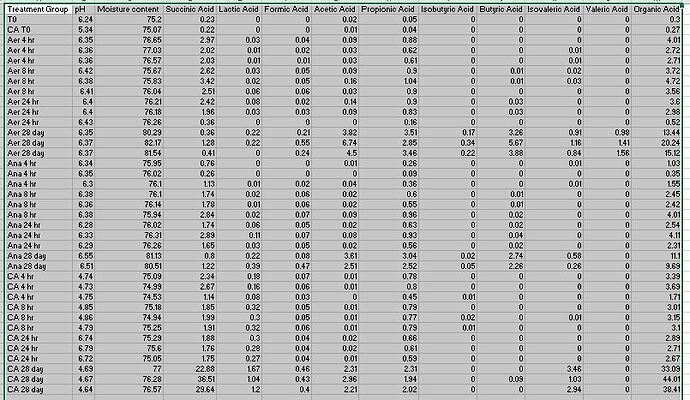
organic_acids<-read.csv(file='OCT19_Queue_organic acid summary8.csv')
organic_acids
summary(organic_acids)
# FORMATTING SPREADSHEET COLUMN NAMES AND TYPES ###############################
colnames(organic_acids)<-gsub("X..","",colnames(organic_acids))
colnames(organic_acids)<-gsub("..db.","",colnames(organic_acids))
colnames(organic_acids)<-gsub(".Acid","_Acid",colnames(organic_acids))
colnames(organic_acids)<-gsub("ï..","",colnames(organic_acids))
organic_acids<-organic_acids %>%
rename("experiment_id"='Experiment.ID',
'sample_name'='OA.sample.name',
'treatment_group'='Sample_overview',
'wet_biomass_extracted_g'='Wet.Biomass.extracted',
'mc_content'='Moisture.content',
'dry_biomass_g'='Dry.Biomass.extracted')
organic_acids<-organic_acids %>%
mutate(across(contains(c("mc_content","MC_avg","_Acid","A_avg","total_avg","A_stdev","total_stdev")),parse_number))
organic_acids
# summary(organic_acids)
# view(organic_acids)
# sapply(organic_acids,class)
# CREATING DATA PLOTS #########################################################
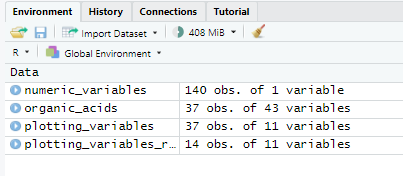
plotting_variables<-organic_acids %>%
select(treatment_group,SA_avg,LA_avg,FA_avg,AA_avg,PA_avg,IBA_avg,BA_avg,IVA_avg,VA_avg,
OA_total_avg)
plotting_variables_remove_na<-na.omit(plotting_variables)
plotting_variables_remove_na
numeric_variables<-plotting_variables_remove_na %>%
group_by(SA_avg,LA_avg,FA_avg,AA_avg,PA_avg,IBA_avg,BA_avg,IVA_avg,VA_avg,
OA_total_avg)
numeric_variables
ggplot(plotting_variables_remove_na,aes(fill=numeric_variables,
y=numeric_variables,x=treatment_group))+
geom_bar()
Output:
> plotting_variables<-organic_acids %>%
+ select(treatment_group,SA_avg,LA_avg,FA_avg,AA_avg,PA_avg,IBA_avg,BA_avg,IVA_avg,VA_avg,
+ OA_total_avg)
> plotting_variables_remove_na<-na.omit(plotting_variables)
> plotting_variables_remove_na
treatment_group SA_avg LA_avg FA_avg AA_avg PA_avg IBA_avg BA_avg IVA_avg VA_avg OA_total_avg
1 Initial biomass 0.23 0.00 0.00 0.02 0.05 0.00 0.00 0.00 0.00 0.30
2 Initial biomass with CA 0.22 0.00 0.00 0.01 0.04 0.00 0.00 0.00 0.00 0.27
3 Aer_4hr 2.34 0.02 0.03 0.05 0.70 0.00 0.00 0.01 0.00 3.15
6 Aer_8hr 2.85 0.04 0.05 0.10 0.95 0.00 0.01 0.02 0.00 4.00
9 Aer_24hr 1.58 0.04 0.02 0.08 0.63 0.00 0.02 0.00 0.00 2.37
12 Aer_28day 0.68 0.15 0.34 5.02 3.27 0.25 4.27 0.97 1.32 16.27
15 Ana_4hr 0.71 0.00 0.01 0.02 0.23 0.00 0.00 0.00 0.00 0.98
18 Ana_8hr 2.12 0.02 0.06 0.04 0.70 0.00 0.01 0.00 0.00 2.96
21 Ana_24hr 2.10 0.07 0.06 0.04 0.71 0.00 0.03 0.00 0.00 2.99
24 Ana_28day 1.01 0.30 0.27 3.06 2.78 0.04 2.50 0.42 0.00 10.39
26 CA_4hr 2.05 0.14 0.05 0.01 0.68 0.00 0.00 0.00 0.00 2.93
29 CA_8hr 1.92 0.31 0.06 0.01 0.78 0.01 0.00 0.00 0.00 3.09
32 CA_24hr 1.79 0.28 0.04 0.02 0.62 0.00 0.00 0.00 0.00 2.76
35 CA_28day 29.68 1.30 0.43 2.49 2.09 0.00 0.03 2.48 0.00 38.50
> numeric_variables<-plotting_variables_remove_na %>%
+ group_by(SA_avg,LA_avg,FA_avg,AA_avg,PA_avg,IBA_avg,BA_avg,IVA_avg,VA_avg,
+ OA_total_avg)
> numeric_variables
# A tibble: 14 x 11
# Groups: SA_avg, LA_avg, FA_avg, AA_avg, PA_avg, IBA_avg, BA_avg, IVA_avg, VA_avg, OA_total_avg [14]
treatment_group SA_avg LA_avg FA_avg AA_avg PA_avg IBA_avg BA_avg IVA_avg VA_avg OA_total_avg
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Initial biomass 0.23 0 0 0.02 0.05 0 0 0 0 0.3
2 Initial biomass with CA 0.22 0 0 0.01 0.04 0 0 0 0 0.27
3 Aer_4hr 2.34 0.02 0.03 0.05 0.7 0 0 0.01 0 3.15
4 Aer_8hr 2.85 0.04 0.05 0.1 0.95 0 0.01 0.02 0 4
5 Aer_24hr 1.58 0.04 0.02 0.08 0.63 0 0.02 0 0 2.37
6 Aer_28day 0.68 0.15 0.34 5.02 3.27 0.25 4.27 0.97 1.32 16.3
7 Ana_4hr 0.71 0 0.01 0.02 0.23 0 0 0 0 0.98
8 Ana_8hr 2.12 0.02 0.06 0.04 0.7 0 0.01 0 0 2.96
9 Ana_24hr 2.1 0.07 0.06 0.04 0.71 0 0.03 0 0 2.99
10 Ana_28day 1.01 0.3 0.27 3.06 2.78 0.04 2.5 0.42 0 10.4
11 CA_4hr 2.05 0.14 0.05 0.01 0.68 0 0 0 0 2.93
12 CA_8hr 1.92 0.31 0.06 0.01 0.78 0.01 0 0 0 3.09
13 CA_24hr 1.79 0.28 0.04 0.02 0.62 0 0 0 0 2.76
14 CA_28day 29.7 1.3 0.43 2.49 2.09 0 0.03 2.48 0 38.5
> ggplot(plotting_variables_remove_na,aes(fill=numeric_variables,
+ y=numeric_variables,x=treatment_group))+
+ geom_bar()
Don't know how to automatically pick scale for object of type grouped_df/tbl_df/tbl/data.frame. Defaulting to continuous.
Don't know how to automatically pick scale for object of type grouped_df/tbl_df/tbl/data.frame. Defaulting to continuous.
Error in `check_aesthetics()`:
! Aesthetics must be either length 1 or the same as the data (14): y and fill
Run `rlang::last_error()` to see where the error occurred.
I've also tried typing in the different columns as the y= and fill= as below:
code:
ggplot(plotting_variables_remove_na,aes(fill=c(SA_avg,LA_avg,AA_avg,PA_avg,
IBA_avg,BA_avg,IVA_avg,VA_avg,
OA_total_avg),
y=c(SA_avg,LA_avg,AA_avg,PA_avg,IBA_avg,
BA_avg,IVA_avg,VA_avg,
OA_total_avg),x=treatment_group))+
geom_bar()
output:
Error in `check_aesthetics()`:
! Aesthetics must be either length 1 or the same as the data (14): y and fill
Run `rlang::last_error()` to see where the error occurred.