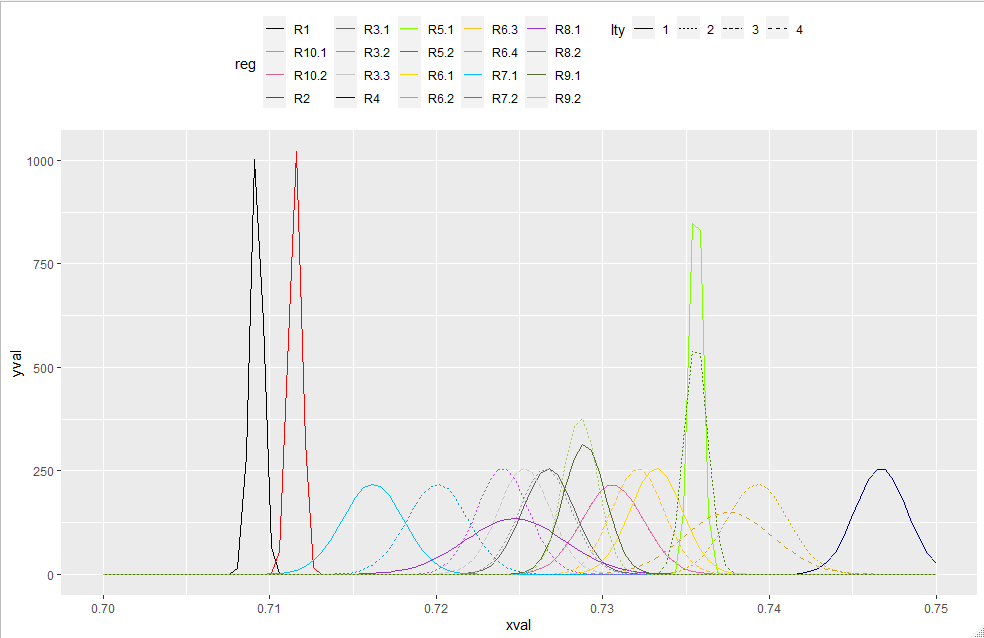
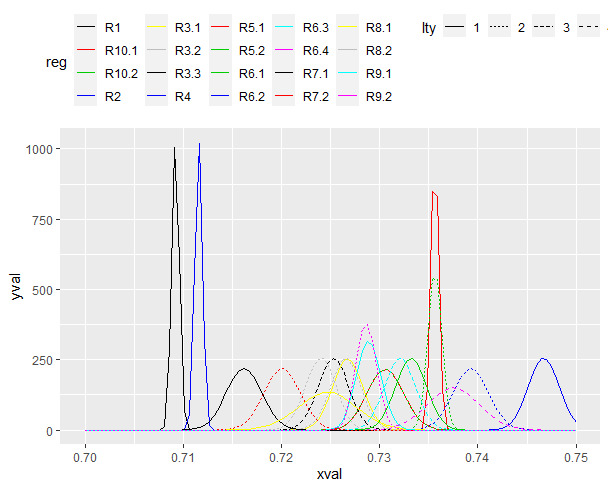
I have created a plot with mulitple normal distributions using the mapply and stat_fuction function in ggplot. I have assigned each line a unique color and line type (dashed or solid), but I have not figured out how to include a legend in the figure. This is my first time using the stat_function in ggplot and an completely lost on how this function talks with the aesthetic mapping funtion (aes) to create the legend. I need the legend to show both the color and line type (dashed or solid) for each normal distribution. The code below will produce the figure without the legend, but has my attempt at producing the legend.
Any assitance would be greatly appreciated. Thanks in advance.
library(ggplot2)
p1 <- ggplot(data = data.frame(x = c(0.707, 0.757)), aes(x = x)) +
mapply(function(mean, sd, col, lty, reg) {
stat_function(fun = dnorm, args = list(mean = mean, sd = sd), aes(colour = reg), col = col, lty = lty, lwd = 1.5)
},
# mean, sd, col, lty
mean = c(0.7092, 0.711533333, 0.72675, 0.726566667, 0.7253, 0.7467, 0.7356, 0.7356, 0.7332, 0.7393, 0.7321, 0.737533333, 0.7162, 0.7201, 0.7247,
0.724, 0.7289, 0.728657143, 0.7306, 0.7306),
sd = c(0.000381708, 0.000381708, 0.001557951, 0.001557951, 0.001557951, 0.001557951, 0.000381708, 0.000696549, 0.001557951, 0.001827967,
0.001557951, 0.002658488, 0.001827967, 0.001827967, 0.002968083, 0.001557951, 0.001266277, 0.001051011, 0.001842546, 0.001842546),
reg = c("R1", "R2", "R3.1", "R3.2", "R3.3", "R4", "R5.1", "R5.2", "R6.1", "R6.2", "R6.3", "R6.4",
"R7.1", "R7.2", "R8.1", "R8.2", "R9.1", "R9.2", "R10.1", "R10.2"),
col = c("black", "red", "gray38", "gray58", "gray78", "darkblue", "chartreuse", "chartreuse4", "gold", "gold3", "goldenrod1", "goldenrod3",
"deepskyblue", "deepskyblue3", "darkorchid", "darkorchid1", "darkolivegreen", "darkolivegreen3", "hotpink", "hotpink3"),
lty = c(1, 1, 1, 2, 3, 1, 1, 2, 1, 2, 3, 4, 1, 2, 1, 2, 1, 2, 1, 2))
p1 + scale_colour_manual(name = "Regions",
values = c("black", "red", "gray38", "gray58", "gray78", "darkblue", "chartreuse", "chartreuse4", "gold", "gold3", "goldenrod1", "goldenrod3",
"deepskyblue", "deepskyblue3", "darkorchid", "darkorchid1", "darkolivegreen", "darkolivegreen3", "hotpink", "hotpink3"),
breaks = c("R1", "R2", "R3.1", "R3.2", "R3.3", "R4", "R5.1", "R5.2", "R6.1", "R6.2", "R6.3", "R6.4",
"R7.1", "R7.2", "R8.1", "R8.2", "R9.1", "R9.2", "R10.1", "R10.2"),
labels = c("1", "2", "3.1", "3.2", "3.3", "4", "5.1", "5.2", "6.1", "6.2", "6.3", "6.4",
"7.1", "7.2", "8.1", "8.2", "9.1", "9.2", "10.1", "10.2"))
p1 + scale_linetype_manual(values = c(1, 1, 1, 2, 3, 1, 1, 2, 1, 2, 3, 4, 1, 2, 1, 2, 1, 2, 1, 2))
p1 + theme(legend.position="top")
p1