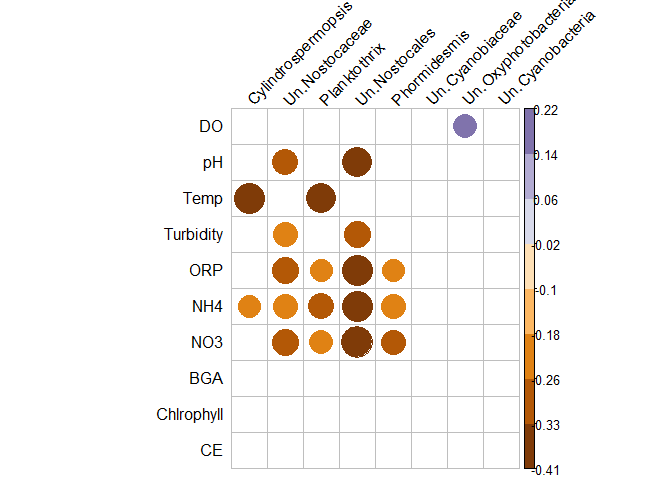
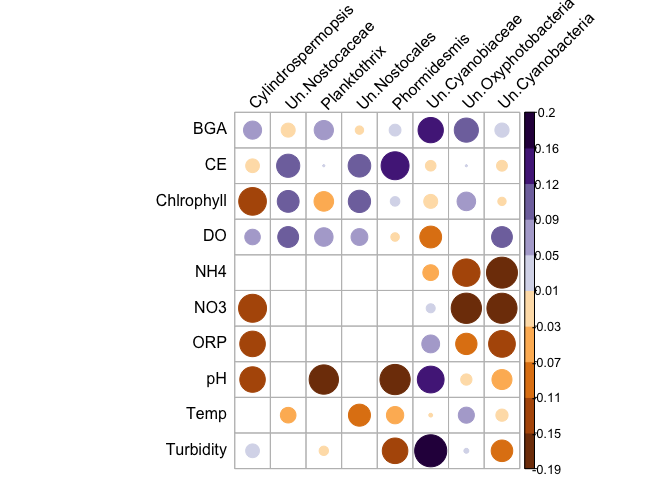
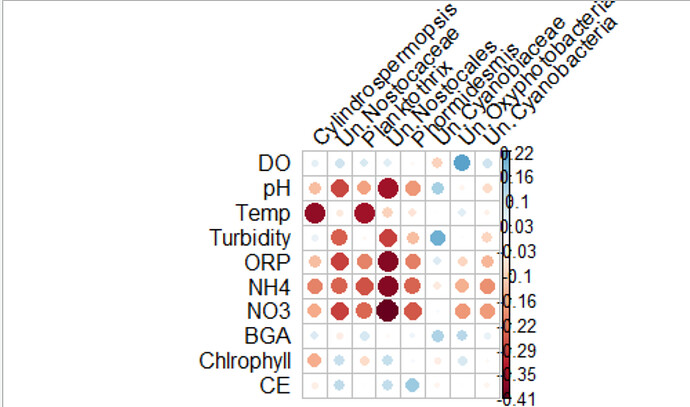
Hello everyone, I am having problems with the cor.mtest function. Is it possible to obtain the level of significance of two different set of variables, and then eliminate those that are below 0.05 (p-value) from the correlation graph?
This is what I tried to do.
library(tidyverse)
library(corrplot)
#> corrplot 0.84 loaded
library(corrr)
metadata <- data.frame(tibble::tribble(
~SampleID, ~DO, ~pH, ~Temp, ~Turbidity, ~ORP, ~NH4, ~NO3, ~BGA, ~Chlrophyll, ~CE,
"1A", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074,
"1B", 2.58, 10.06, 24.95, 95.2, 108.4, 4.62, 3.98, 279113, 38.9, 1.082,
"1C", 2.85, 9.98, 24.01, 98.8, 102.9, 4.37, 3.57, 265304, 40, 1.081,
"1D", 3.81, 9.07, 26.67, 24.9, 94, 5.6, 3.53, 11603, 44, 1.147,
"1E", 2.64, 8.99, 24.01, 43.1, 63.9, 3.38, 2.55, 218471, 35, 1.156,
"1F", 4.61, 8.93, 23.5, 49.1, 64.7, 3.21, 2.28, 226658, 32.8, 1.157,
"1G", 5.74, 9.24, 24.46, 68.6, 40.3, 1.93, 0.71, 267389, 44, 1.072,
"1H", 3.69, 9.32, 24.65, 69.8, 48.1, 2.21, 1.2, 267144, 44.8, 1.152,
"2A", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074,
"2B", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081,
"2C", 2.85, 9.98, 24.01, 98.8, 102.9, 4.37, 3.57, 265304, 40, 1.081,
"2D", 3.81, 9.07, 26.67, 24.9, 94, 5.6, 3.53, 11603, 44, 1.147,
"2E", 2.64, 8.99, 24.01, 43.1, 63.9, 3.38, 2.55, 218471, 35, 1.156,
"2F", 4.61, 8.93, 23.5, 49.1, 64.7, 3.21, 2.28, 226658, 32.8, 1.157,
"2G", 5.74, 9.24, 24.46, 68.6, 40.3, 1.93, 0.71, 267389, 44, 1.072,
"2H", 3.69, 9.32, 24.65, 69.8, 48.1, 2.21, 1.2, 267144, 44.8, 1.152,
"3A", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075,
"3B", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081,
"3C", 3.14, 10, 24, 96.9, 108, 4.48, 3.76, 279119, 38.5, 1.08,
"3D", 2.92, 9.06, 26.46, 39.7, 23.7, 7.64, 3.66, 66608, 45.1, 1.138,
"3E", 1.83, 8.99, 23.99, 49, 64.1, 3.18, 2.44, 213497, 42.2, 1.155,
"3F", 1.82, 8.92, 23.43, 47.6, 60.4, 3.09, 2.06, 242331, 50.2, 1.157,
"3G", 3.02, 9.18, 24.14, 66.6, 40.6, 2.19, 0.67, 267007, 43.6, 1.061,
"4A", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075,
"4B", 3.04, 9.96, 24.04, 98.5, 98.9, 3.97, 4.35, 279109, 43, 1.083,
"4C", 2.85, 9.98, 24.01, 98.8, 102.9, 4.37, 3.57, 265304, 40, 1.081,
"4D", 2.92, 9.06, 26.46, 39.7, 23.7, 7.64, 3.66, 66608, 45.1, 1.138,
"4E", 1.83, 8.99, 23.99, 49, 64.1, 3.18, 2.44, 213497, 42.2, 1.155,
"4F", 1.82, 8.92, 23.43, 47.6, 60.4, 3.09, 2.06, 242331, 50.2, 1.157,
"4G", 3.02, 9.18, 24.14, 66.6, 40.6, 2.19, 0.67, 267007, 43.6, 1.061,
"4H", 3.3, 9.32, 24.67, 74.1, 49.8, 2.14, 0.87, 261363, 45.1, 1.142,
"5A", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074,
"5B", 3.04, 9.96, 24.04, 98.5, 98.9, 3.97, 4.35, 279109, 43, 1.083,
"5D", 3.81, 9.07, 26.67, 24.9, 94, 5.6, 3.53, 11603, 44, 1.147,
"5E", 2.64, 8.99, 24.01, 43.1, 63.9, 3.38, 2.55, 218471, 35, 1.156,
"5F", 4.61, 8.93, 23.5, 49.1, 64.7, 3.21, 2.28, 226658, 32.8, 1.157,
"5G", 5.74, 9.24, 24.46, 68.6, 40.3, 1.93, 0.71, 267389, 44, 1.072,
"5H", 3.69, 9.32, 24.65, 69.8, 48.1, 2.21, 1.2, 267144, 44.8, 1.152,
"6A", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074,
"6B", 2.09, 9.95, 24.01, 95.9, 92.8, 4.06, 3.68, 279100, 50, 1.077,
"6C", 2.98, 10.09, 24.36, 97.2, 106.9, 4.43, 3.34, 279099, 43, 1.077,
"6D", 3.81, 9.07, 26.67, 24.9, 94, 5.6, 3.53, 11603, 44, 1.147,
"6E", 1.83, 8.99, 23.99, 49, 64.1, 3.18, 2.44, 213497, 42.2, 1.155,
"6F", 1.82, 8.92, 23.43, 47.6, 60.4, 3.09, 2.06, 242331, 50.2, 1.157,
"6G", 3.02, 9.18, 24.14, 66.6, 40.6, 2.19, 0.67, 267007, 43.6, 1.061,
"6H", 3.3, 9.32, 24.67, 74.1, 49.8, 2.14, 0.87, 261363, 45.1, 1.142,
"7A", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075,
"7B", 2.09, 9.95, 24.01, 95.9, 92.8, 4.06, 3.68, 279100, 50, 1.081,
"7C", 2.98, 10.09, 24.36, 97.2, 106.9, 4.43, 3.34, 279099, 43, 1.077,
"7D", 8.17, 9.04, 24.88, 46.9, 90.4, 2.81, 2.25, 246979, 34, 1.154,
"7E", 2.63, 9, 24.7, 37.2, 74.9, 4.2, 2.99, 169175, 38.4, 1.179,
"7F", 7.69, 9.53, 26.14, 74.2, 67.7, 4.5, 1.3, 272332, 58.5, 1.072,
"7G", 3.07, 9.31, 24.19, 69.9, 42, 1.99, 0.74, 269519, 44.3, 1.152,
"7H", 3.7, 9.14, 22.65, 71.3, 37.1, 2.07, 0.68, 272096, 40, 1.127,
"8A", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075,
"8B", 3.04, 9.96, 24.04, 98.5, 98.9, 3.97, 4.35, 279109, 43, 1.083,
"8C", 2.18, 9.97, 24.4, 100.6, 102.7, 5.47, 4.11, 278137, 53.8, 1.078,
"8D", 8.17, 9.04, 24.88, 46.9, 90.4, 2.81, 2.25, 246979, 34, 1.154,
"8E", 2.63, 9, 24.7, 37.2, 74.9, 4.2, 2.99, 169175, 38.4, 1.179,
"8F", 7.69, 9.53, 26.14, 74.2, 67.7, 4.5, 1.3, 272332, 58.5, 1.072,
"8G", 3.07, 9.31, 24.19, 69.9, 42, 1.99, 0.74, 269519, 44.3, 1.152,
"8H", 3.7, 9.14, 22.65, 71.3, 37.1, 2.07, 0.68, 272096, 40, 1.127,
"9A", 2.58, 10.06, 24.95, 95.2, 108.4, 4.62, 3.98, 279113, 38.9, 1.082,
"9B", 2.09, 9.95, 24.01, 95.9, 92.8, 4.06, 3.68, 279100, 50, 1.081,
"9C", 2.18, 9.97, 24.4, 100.6, 102.7, 5.47, 4.11, 278137, 53.8, 1.078,
"9D", 2.16, 8.96, 24.3, 42, 53, 3.19, 2.1, 174271, 50.3, 1.161,
"9E", 1.7, 9.02, 24.72, 36.1, 63.4, 3.97, 2.55, 159046, 55.9, 1.69,
"9F", 6.8, 9.51, 26.15, 74.4, 73.9, 3.3, 1.11, 271503, 51.3, 1.061,
"9G", 1.95, 9.39, 23.9, 44.4, 42, 3.38, 0.69, 261124, 68.2, 1.147,
"9H", 4.28, 8.85, 20.65, 71.3, 37.5, 1.18, 0.69, 271303, 34.2, 1.102,
"10A", 2.58, 10.06, 24.95, 95.2, 108.4, 4.62, 3.98, 279113, 38.9, 1.082,
"10B", 2.09, 9.95, 24.01, 95.9, 92.8, 4.06, 3.68, 279100, 50, 1.081,
"10C", 2.98, 10.09, 24.36, 97.2, 106.9, 4.43, 3.34, 279099, 43, 1.077,
"10D", 2.16, 8.96, 24.3, 42, 53, 3.19, 2.1, 174271, 50.3, 1.161,
"10E", 1.7, 9.02, 24.72, 36.1, 63.4, 3.97, 2.55, 159046, 55.9, 1.69,
"10F", 6.8, 9.51, 26.15, 74.4, 73.9, 3.3, 1.11, 271503, 51.3, 1.061,
"10G", 1.95, 9.39, 23.9, 44.4, 42, 3.38, 0.69, 261124, 68.2, 1.147,
"10H", 4.28, 8.85, 20.65, 71.3, 37.5, 1.18, 0.69, 271303, 34.2, 1.102,
"11A", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081,
"11B", 3.14, 10, 24, 96.9, 108, 4.48, 3.76, 279119, 38.5, 1.08,
"11C", 2.18, 9.97, 24.4, 100.6, 102.7, 5.47, 4.11, 278137, 53.8, 1.078,
"11D", 8.17, 9.04, 24.88, 46.9, 90.4, 2.81, 2.25, 246979, 34, 1.154,
"11E", 2.63, 9, 24.7, 37.2, 74.9, 4.2, 2.99, 169175, 38.4, 1.179,
"11F", 7.69, 9.53, 26.14, 74.2, 67.7, 4.5, 1.3, 272332, 58.5, 1.072,
"11G", 3.07, 9.31, 24.19, 69.9, 42, 1.99, 0.74, 269519, 44.3, 1.152,
"11H", 3.7, 9.14, 22.65, 71.3, 37.1, 2.07, 0.68, 272096, 40, 1.127,
"12A", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081,
"12B", 3.14, 10, 24, 96.9, 111.9, 4.48, 3.76, 279119, 38.5, 1.08,
"12C", 2.92, 9.06, 26.46, 39.7, 23.7, 7.64, 3.66, 66608, 45.1, 1.138,
"12D", 2.16, 8.96, 24.3, 42, 53, 3.19, 2.1, 174271, 50.3, 1.161,
"12E", 1.7, 9.02, 24.72, 36.1, 63.4, 3.97, 2.55, 159046, 55.9, 1.69,
"12F", 6.8, 9.51, 26.15, 74.4, 73.9, 3.3, 1.11, 271503, 51.3, 1.061,
"12G", 1.95, 9.39, 23.9, 44.4, 42, 3.38, 0.69, 261124, 68.2, 1.147,
"12H", 4.28, 8.85, 20.65, 71.3, 37.5, 1.18, 0.69, 271303, 34.2, 1.102
)
)
bacteria <- data.frame (tibble::tribble(
~index, ~Cylindrospermopsis, ~Un.Nostocaceae, ~Planktothrix, ~Un.Nostocales, ~Phormidesmis, ~Un.Cyanobiaceae, ~Un.Oxyphotobacteria, ~Un.Cyanobacteria,
"1A", 0, 0, 0, 12, 66, 0, 7137, 1467,
"1B", 0, 0, 0, 6, 35, 0, 5549, 238,
"1C", 0, 19, 2, 0, 96, 0, 1861, 708,
"1D", 0, 0, 0, 0, 66, 19, 3185, 4001,
"1E", 0, 1662, 0, 207, 720, 529, 7468, 1928,
"1F", 0, 2090, 0, 425, 432, 6, 18421, 4460,
"1G", 0, 372, 0, 0, 19, 2, 4191, 361,
"1H", 0, 433, 613, 46, 1215, 0, 12240, 1970,
"2A", 0, 1000, 0, 8, 195, 0, 20728, 4350,
"2B", 0, 769, 35, 62, 46, 619, 13438, 3076,
"2C", 0, 639, 0, 31, 14, 21, 17664, 2008,
"2D", 0, 510, 0, 0, 58, 0, 5085, 2745,
"2E", 0, 319, 0, 0, 60, 200, 8376, 1613,
"2F", 0, 501, 2, 89, 807, 0, 8906, 983,
"2G", 0, 930, 0, 115, 141, 195, 13273, 1514,
"2H", 9, 1867, 18, 535, 279, 26, 18506, 3531,
"3A", 44, 272, 88, 3, 1139, 0, 7959, 1700,
"3B", 570, 43, 60, 22, 65, 3, 762, 367,
"3C", 0, 0, 0, 0, 173, 0, 3088, 45,
"3D", 0, 0, 0, 0, 0, 52, 3009, 1683,
"3E", 0, 1039, 0, 199, 74, 0, 4120, 754,
"3F", 0, 388, 0, 10, 318, 0, 5083, 675,
"3G", 0, 982, 0, 298, 407, 0, 5573, 1031,
"4A", 0, 3272, 0, 578, 558, 32, 24267, 9707,
"4B", 0, 895, 0, 60, 33, 1685, 16403, 3105,
"4C", 0, 243, 0, 0, 0, 57, 6043, 570,
"4D", 0, 1621, 0, 148, 15, 6, 8472, 2357,
"4E", 0, 1094, 0, 168, 165, 0, 8351, 2424,
"4F", 0, 43, 0, 0, 302, 0, 491, 570,
"4G", 0, 291, 0, 74, 152, 359, 13031, 3066,
"4H", 16, 2814, 28, 616, 1841, 0, 33094, 6509,
"5A", 0, 2, 0, 111, 2, 58, 11083, 4240,
"5B", 0, 0, 0, 0, 108, 0, 4248, 676,
"5D", 0, 1006, 0, 214, 285, 0, 8839, 1176,
"5E", 0, 2, 17, 120, 355, 19, 10286, 213,
"5F", 0, 1051, 2, 158, 583, 0, 14778, 1765,
"5G", 2, 1040, 28, 587, 9, 355, 3584, 1574,
"5H", 0, 244, 0, 70, 55, 22, 10905, 7012,
"6A", 37, 1221, 0, 126, 43, 473, 22140, 5237,
"6B", 0, 1190, 0, 92, 140, 599, 22958, 3646,
"6C", 0, 266, 0, 0, 46, 0, 5937, 1510,
"6D", 0, 63, 0, 7, 0, 0, 8240, 900,
"6E", 0, 1016, 0, 201, 130, 0, 6252, 1583,
"6F", 0, 2226, 0, 491, 190, 0, 15494, 3392,
"6G", 0, 206, 0, 296, 8, 205, 9631, 5514,
"6H", 0, 2014, 123, 430, 3015, 74, 40839, 7004,
"7A", 0, 183, 0, 16, 172, 50, 7201, 4795,
"7B", 0, 0, 0, 0, 0, 0, 2146, 732,
"7C", 0, 27, 0, 0, 0, 0, 3927, 450,
"7D", 0, 139, 128, 42, 97, 11, 26197, 1932,
"7E", 0, 1266, 0, 274, 1208, 0, 17161, 4310,
"7F", 0, 232, 274, 3, 335, 60, 14640, 293,
"7G", 0, 1036, 0, 210, 5, 424, 1888, 1837,
"7H", 0, 102, 0, 0, 0, 0, 2860, 2943,
"8A", 0, 1096, 0, 83, 56, 16, 18949, 3364,
"8B", 0, 2439, 0, 263, 117, 48, 30505, 6066,
"8C", 0, 39, 29, 0, 102, 0, 10406, 572,
"8D", 0, 2214, 0, 324, 97, 0, 14814, 8608,
"8E", 0, 1709, 102, 323, 6, 0, 7553, 2455,
"8F", 0, 2633, 0, 237, 391, 0, 24075, 3692,
"8G", 45, 2557, 442, 610, 108, 1817, 7788, 4399,
"8H", 0, 1403, 24, 251, 256, 4, 16103, 3045,
"9A", 0, 2, 0, 20, 6, 0, 3794, 1919,
"9B", 0, 5, 0, 30, 483, 8, 8916, 1307,
"9C", 0, 46, 0, 0, 0, 0, 2817, 1041,
"9D", 0, 3157, 0, 706, 162, 0, 17692, 8381,
"9E", 0, 342, 125, 64, 1240, 380, 20302, 2008,
"9F", 8, 1023, 0, 257, 584, 12, 48687, 5115,
"9G", 0, 407, 0, 72, 75, 195, 14834, 2512,
"9H", 3395, 2082, 2796, 357, 540, 0, 4402, 1391,
"10A", 0, 654, 0, 37, 116, 120, 14300, 3054,
"10B", 0, 343, 0, 0, 0, 77, 15485, 1203,
"10C", 0, 358, 0, 0, 0, 0, 2695, 1719,
"10D", 0, 38, 47, 0, 18, 37, 13401, 342,
"10E", 0, 2265, 0, 382, 28, 15, 9052, 2779,
"10F", 0, 824, 0, 127, 76, 0, 13282, 743,
"10G", 0, 338, 40, 5, 281, 13, 2492, 939,
"10H", 0, 151, 0, 0, 37, 0, 2924, 700,
"11A", 0, 11, 0, 0, 6, 2111, 317, 1532,
"11B", 0, 27, 0, 0, 0, 0, 968, 702,
"11C", 0, 0, 0, 0, 62, 9, 4148, 2733,
"11D", 0, 1108, 0, 211, 165, 0, 10756, 4030,
"11E", 0, 1105, 0, 190, 304, 0, 10605, 1595,
"11F", 0, 0, 0, 19, 70, 20, 4327, 390,
"11G", 0, 0, 0, 16, 54, 56, 6701, 2077,
"11H", 0, 88, 0, 0, 20, 0, 14377, 1305,
"12A", 0, 441, 0, 50, 123, 115, 29898, 2216,
"12B", 0, 404, 0, 0, 59, 55, 11845, 1397,
"12C", 0, 623, 0, 6, 57, 0, 18824, 1992,
"12D", 77, 3141, 145, 695, 172, 166, 9555, 5101,
"12E", 0, 172, 0, 0, 67, 5, 6934, 243,
"12F", 0, 2631, 0, 425, 74, 11, 16460, 2880,
"12G", 2, 3522, 319, 784, 672, 0, 29848, 9140,
"12H", 280, 1552, 216, 350, 551, 10, 33984, 7351
)
)
library(RColorBrewer)
res1 <- cor.mtest(bind_cols(bacteria[, -1],metadata[, -1]), conf.level = 0.95)
corrr::correlate(bind_cols(metadata[, -1], bacteria[, -1])) %>%
filter(rowname %in% colnames(metadata)) %>%
select(one_of(c("rowname", colnames(bacteria)[-1]))) %>%
as.data.frame() %>%
column_to_rownames("rowname") %>%
as.matrix() %>%
corrplot::corrplot(is.corr = FALSE, p.mat = res1$p, insig= "blank", sig.level = 0.05, tl.col = "black", tl.srt = 45,
col= brewer.pal (n=10, name= "PuOr"))
#>
#> Correlation method: 'pearson'
#> Missing treated using: 'pairwise.complete.obs'

Created on 2019-11-04 by the reprex package (v0.3.0)
The graphic is wrong.
The example I found was this: An Introduction to corrplot Package
Any help in getting a complete set of information would be greatly appreciated!
Greetings,
Osiris