JRH
December 20, 2023, 8:08pm
1
Hello, I have a column C that has rows each containing unique strings. I would like to copy one part of a string, the protein ID, to parts of the string that do not have that allocated, if applicable. You can see that most of the [ranges] in the brackets have an ID before them, but in C1 and C2 below there is each a set that does not. I want the previous ID to be placed there. Here is the example. Thanks.
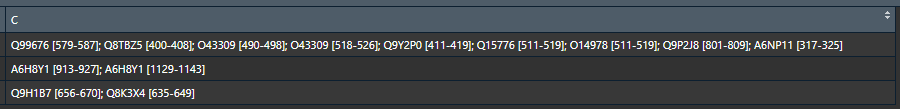
C1 <- c("Q99676 [579-587]; Q8TBZ5 [400-408]; O43309 [490-498]; [518-526]; Q9Y2P0 [411-419]; Q15776 [511-519]; O14978 [511-519]; Q9P2J8 [801-809]; A6NP11 [317-325]")
C2 <- c("A6H8Y1 [913-927]; [1129-1143]")
c3<- c("Q9H1B7 [656-670]; Q8K3X4 [635-649]")
## I would like the output to be
C1 <- c("Q99676 [579-587]; Q8TBZ5 [400-408]; O43309 [490-498]; O43309 [518-526]; Q9Y2P0 [411-419]; Q15776 [511-519]; O14978 [511-519]; Q9P2J8 [801-809]; A6NP11 [317-325]")
C2 <- c("A6H8Y1 [913-927]; A6H8Y1 [1129-1143]")
c3<- c("Q9H1B7 [656-670]; Q8K3X4 [635-649]")
AlexisW
December 20, 2023, 9:24pm
2
I can suggest a tidyverse approach:
C1 |>
str_split_1(";") |>
str_trim() |>
enframe() |>
separate_wider_delim(value,
delim = " ",
names = c("protein_ID", "range"),
too_few = "align_end") |>
fill(protein_ID)
first, make this single long string into a vector, and enframe() it to make it into a data frame column. Then you can split the protein ID from the range, and simply use fill() to fill up the missing values.
1 Like
Hi @JRH . Below is one approach that achieves the desired output.
C1 <- c("Q99676 [579-587]; Q8TBZ5 [400-408]; O43309 [490-498]; [518-526]; Q9Y2P0 [411-419]; Q15776 [511-519]; O14978 [511-519]; Q9P2J8 [801-809]; A6NP11 [317-325]")
C2 <- c("A6H8Y1 [913-927]; [1129-1143]")
C3 <- c("Q9H1B7 [656-670]; Q8K3X4 [635-649]")
df = data.frame(C = c(C1, C2, C3))
library(tidyverse)
df |>
mutate(row = row_number()) |>
mutate(C = str_replace_all(C, '; ', '|')) |>
separate_rows(C, sep = '\\|') |>
mutate(C = str_trim(C)) |>
separate(C, sep = ' ', into = c('id', 'range')) |>
mutate(range = ifelse(is.na(range), id, range)) |>
group_by(row) |>
mutate(id = ifelse(id == range, lag(id), id)) |>
mutate(C = paste(id, range)) |>
mutate(C = paste(C, collapse = '; ')) |>
ungroup() |>
distinct(C)
1 Like
AlexisW
December 20, 2023, 11:07pm
4
Oh and if you didn't want to go through a data.frame:
library(stringr)
C1 <- c("Q99676 [579-587]; Q8TBZ5 [400-408]; O43309 [490-498]; [518-526]; Q9Y2P0 [411-419]; Q15776 [511-519]; O14978 [511-519]; Q9P2J8 [801-809]; A6NP11 [317-325]")
C2 <- c("A6H8Y1 [913-927]; [1129-1143]")
c3<- c("Q9H1B7 [656-670]; Q8K3X4 [635-649]")
fill_vector <- function(x){
for(i in seq_along(x)[-1]){
if(is.na(x[[i]])) x[[i]] <- x[[i-1]]
}
x
}
C1 |>
str_split_1(";") |>
str_trim() |>
str_extract("^[A-Z][A-Z0-9]+ ") |>
fill_vector()
#> [1] "Q99676 " "Q8TBZ5 " "O43309 " "O43309 " "Q9Y2P0 " "Q15776 " "O14978 "
#> [8] "Q9P2J8 " "A6NP11 "
C2 |>
str_split_1(";") |>
str_trim() |>
str_extract("^[A-Z][A-Z0-9]+ ") |>
fill_vector()
#> [1] "A6H8Y1 " "A6H8Y1 "
Created on 2023-12-20 with reprex v2.0.2
1 Like
system
December 27, 2023, 11:08pm
5
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.