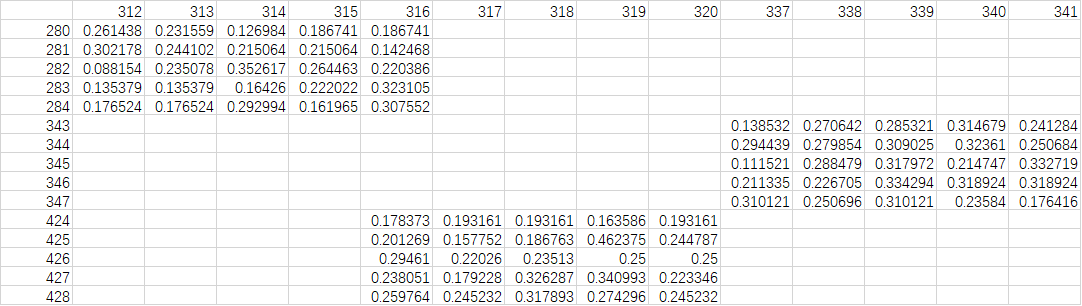
Hi, I have obtain the hyperspectral data (see attachment, pixel 15*5 with 256 channels) from thousand figures using envi software. I am trying to use keras to do deep learning in Rstuio. How can I convert the hyperspectral data to assay, add label, and combined all labled hyperspectral data to build DP models?
9.pdf (600.7 KB)
I have followed the script below, but it's to be wrong.
dataX <- read.csv("9.csv", header =T) #I have save the CSV file to pdf file which was uploaded.
dataX2 <- dataX %>% pivot_longer(cols = c(3:ncol(.)), names_to = "Wavelength", values_to = "Value") %>% mutate(across(everything(), as.numeric))
dataX3 <- dataX2 %>% select(Value) %>% array(dim = c(15, 5, 256))