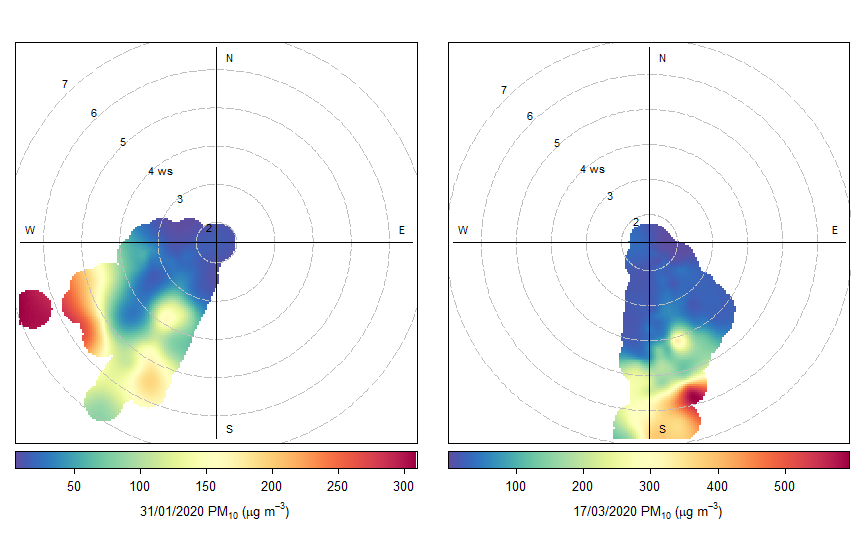
Here's what a reprex looks like. Besides making the problem easier to work with, it helps in catching threshold problems shown below.
library(openair)
# cut and pasted from post; NA added to last field of row 6
# readr::read_csv("~/Desktop/grist.csv") -> Rata_street_data
# produced with dput(Rata_street_data)
Rata_street_data <- structure(list(
date_long =
c("31/01/2020 3:00", "31/01/2020 23:10", "31/01/2020 23:20", "31/01/2020 23:30", "31/01/2020 23:40", "31/01/2020 23:50", "17/03/2020 0:00", "17/03/2020 0:10", "17/03/2020 0:20", "17/03/2020 0:30", "17/03/2020 0:40", "17/03/2020 0:50"),
date =
c("31/01/2020", "31/01/2020", "31/01/2020", "31/01/2020", "31/01/2020", "31/01/2020", "17/03/2020", "17/03/2020", "17/03/2020", "17/03/2020", "17/03/2020", "17/03/2020"), time = structure(c(82800, 83400, 84000, 84600, 85200, 85800, 0, 600, 1200, 1800, 2400, 3000),
class =
c("hms", "difftime"), units = "secs"
),
ws =
c(3.61, 3.262, 2.831, 2.567, 2.648, 2.634, 2.312, 1.925, 2.23, 2.039, 1.74, 2.149),
wd =
c(254.195, 254.037, 260.564, 254.344, 246.959, 253.559, 123.102, 128.463, 132.018, 131.712, 133.473, 126.05),
pm10 =
c(9.489, 5.243, 4.386, 4.062, 3.578, 4.532, 11.375, NA, 13.474, 8.101, -1.888, 2.12)
),
class =
c("spec_tbl_df", "tbl_df", "tbl", "data.frame"),
row.names =
c(NA, -12L), spec = structure(list(cols = list(date_long = structure(list(),
class =
c("collector_character", "collector")
), date = structure(list(),
class =
c("collector_character", "collector")
), time = structure(list(format = ""),
class =
c("collector_time", "collector")
), ws = structure(list(),
class =
c("collector_double", "collector")
), wd = structure(list(),
class =
c("collector_double", "collector")
), pm10 = structure(list(),
class =
c("collector_double", "collector")
)), default = structure(list(),
class =
c("collector_guess", "collector")
), skip = 1L),
class = "col_spec"
)
)
# code and data provided was not used to create the images posted
obj <- polarPlot(selectByDate(Rata_street_data, start = "31/01/2020", end = "31/01/2020"),
pollutant = "pm10",
key.position = "bottom",
key.header = "31/01/2020 PM10 (ug/m3)",
key.footer = NULL
)
#> Warning: Missing dates detected, removing 12 lines
#> Warning in min(mydata[[x]], na.rm = TRUE): no non-missing arguments to min;
#> returning Inf
#> Error in if (length(which(mydata[pollutant] < 0))/nrow(mydata) > 0.1 && : missing value where TRUE/FALSE needed
Using the built-in data, we see
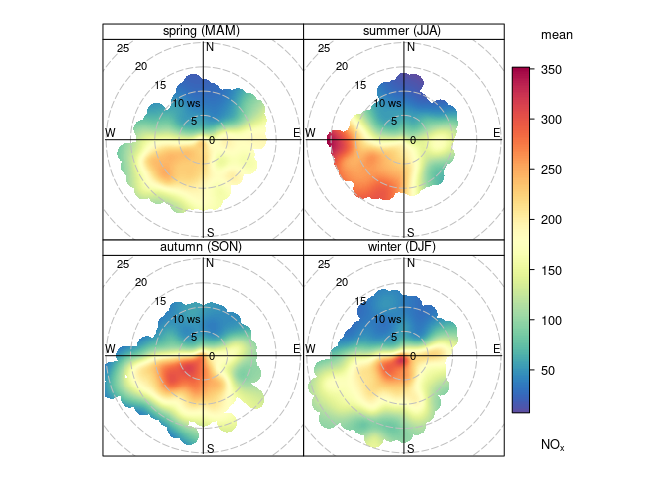
library(openair)
obj <- polarPlot(openair::mydata, pollutant = "nox", type = "season")
str(obj)
#> List of 3
#> $ plot:List of 45
#> ..$ formula :Class 'formula' language z ~ u * v | season
#> .. .. ..- attr(*, ".Environment")=<environment: 0x5588d2e348c8>
#> ..$ as.table : logi TRUE
#> ..$ aspect.fill : logi FALSE
#> ..$ legend :List of 1
#> .. ..$ right:List of 3
#> .. .. ..$ fun :function (key, draw = FALSE, vp = NULL)
#> .. .. ..$ args:List of 1
#> .. .. .. ..$ key:List of 10
#> .. .. .. .. ..$ col : chr [1:199] "#5E4EA2" "#5851A5" "#5253A9" "#4D56AC" ...
#> .. .. .. .. ..$ at : num [1:200] 7.25 8.98 10.71 12.44 14.18 ...
#> .. .. .. .. ..$ labels :List of 2
#> .. .. .. .. .. ..$ labels: num [1:7] 50 100 150 200 250 300 350
#> .. .. .. .. .. ..$ at : num [1:7] 50 100 150 200 250 300 350
#> .. .. .. .. ..$ space : chr "right"
#> .. .. .. .. ..$ auto.text: logi TRUE
#> .. .. .. .. ..$ footer : chr "nox"
#> .. .. .. .. ..$ header : chr "mean"
#> .. .. .. .. ..$ height : num 1
#> .. .. .. .. ..$ width : num 1.5
#> .. .. .. .. ..$ fit : chr "all"
#> .. .. ..$ draw: logi FALSE
#> ..$ panel :function (x, y, z, subscripts, ...)
#> ..$ page : NULL
#> ..$ layout : NULL
#> ..$ skip : logi FALSE
#> ..$ strip :function (...)
#> ..$ strip.left : logi FALSE
#> ..$ xscale.components:function (lim, packet.number = 0, packet.list = NULL, top = TRUE, ...)
#> ..$ yscale.components:function (lim, packet.number = 0, packet.list = NULL, right = TRUE, ...)
#> ..$ axis :function (side = c("top", "bottom", "left", "right"), scales, components,
#> as.table, labels = c("default", "yes", "no"), ticks = c("default",
#> "yes", "no"), ..., prefix = lattice.getStatus("current.prefix"))
#> ..$ xlab : expression(paste())
#> ..$ ylab : expression(paste())
#> ..$ xlab.default : chr "u"
#> ..$ ylab.default : chr "v"
#> ..$ xlab.top : NULL
#> ..$ ylab.right : NULL
#> ..$ main : expression(paste())
#> ..$ sub : NULL
#> ..$ x.between : num 0
#> ..$ y.between : num 0
#> ..$ par.settings : NULL
#> ..$ plot.args : NULL
#> ..$ lattice.options : NULL
#> ..$ par.strip.text :List of 1
#> .. ..$ cex: num 0.8
#> ..$ index.cond :List of 1
#> .. ..$ : int [1:4] 1 2 3 4
#> ..$ perm.cond : int 1
#> ..$ condlevels :List of 1
#> .. ..$ season: chr [1:4] "spring (MAM)" "summer (JJA)" "autumn (SON)" "winter (DJF)"
#> ..$ call : language levelplot(openair::mydata, pollutant = "nox", type = "season")
#> ..$ x.scales :List of 26
#> .. ..$ draw : logi FALSE
#> .. ..$ axs : chr "r"
#> .. ..$ tck : num [1:2] 1 1
#> .. ..$ tick.number : num 5
#> .. ..$ at : logi FALSE
#> .. ..$ labels : logi FALSE
#> .. ..$ log : logi FALSE
#> .. ..$ alternating : num [1:2] 1 2
#> .. ..$ relation : chr "same"
#> .. ..$ abbreviate : logi FALSE
#> .. ..$ minlength : num 4
#> .. ..$ limits : NULL
#> .. ..$ format : NULL
#> .. ..$ equispaced.log: logi TRUE
#> .. ..$ lty : logi FALSE
#> .. ..$ lwd : logi FALSE
#> .. ..$ cex : logi [1:2] FALSE FALSE
#> .. ..$ rot : logi [1:2] FALSE FALSE
#> .. ..$ col : logi FALSE
#> .. ..$ col.line : logi FALSE
#> .. ..$ alpha : logi FALSE
#> .. ..$ alpha.line : logi FALSE
#> .. ..$ font : logi FALSE
#> .. ..$ fontfamily : logi FALSE
#> .. ..$ fontface : logi FALSE
#> .. ..$ lineheight : logi FALSE
#> ..$ y.scales :List of 26
#> .. ..$ draw : logi FALSE
#> .. ..$ axs : chr "r"
#> .. ..$ tck : num [1:2] 1 1
#> .. ..$ tick.number : num 5
#> .. ..$ at : logi FALSE
#> .. ..$ labels : logi FALSE
#> .. ..$ log : logi FALSE
#> .. ..$ alternating : num [1:2] 1 2
#> .. ..$ relation : chr "same"
#> .. ..$ abbreviate : logi FALSE
#> .. ..$ minlength : num 4
#> .. ..$ limits : NULL
#> .. ..$ format : NULL
#> .. ..$ equispaced.log: logi TRUE
#> .. ..$ lty : logi FALSE
#> .. ..$ lwd : logi FALSE
#> .. ..$ cex : logi [1:2] FALSE FALSE
#> .. ..$ rot : logi [1:2] FALSE FALSE
#> .. ..$ col : logi FALSE
#> .. ..$ col.line : logi FALSE
#> .. ..$ alpha : logi FALSE
#> .. ..$ alpha.line : logi FALSE
#> .. ..$ font : logi FALSE
#> .. ..$ fontfamily : logi FALSE
#> .. ..$ fontface : logi FALSE
#> .. ..$ lineheight : logi FALSE
#> ..$ panel.args.common:List of 7
#> .. ..$ x : num [1:161604] -20.2 -20 -19.8 -19.6 -19.4 ...
#> .. ..$ y : num [1:161604] -20.2 -20.2 -20.2 -20.2 -20.2 ...
#> .. ..$ z : num [1:161604] NA NA NA NA NA NA NA NA NA NA ...
#> .. ..$ at : num [1:17] -16.86 7.68 32.23 56.77 81.31 ...
#> .. ..$ region : logi TRUE
#> .. ..$ axes : logi FALSE
#> .. ..$ col.regions: chr [1:199] "#5E4EA2" "#5851A5" "#5253A9" "#4D56AC" ...
#> ..$ panel.args :List of 4
#> .. ..$ :List of 1
#> .. .. ..$ subscripts: int [1:40401] 1 2 3 4 5 6 7 8 9 10 ...
#> .. ..$ :List of 1
#> .. .. ..$ subscripts: int [1:40401] 40402 40403 40404 40405 40406 40407 40408 40409 40410 40411 ...
#> .. ..$ :List of 1
#> .. .. ..$ subscripts: int [1:40401] 80803 80804 80805 80806 80807 80808 80809 80810 80811 80812 ...
#> .. ..$ :List of 1
#> .. .. ..$ subscripts: int [1:40401] 121204 121205 121206 121207 121208 121209 121210 121211 121212 121213 ...
#> ..$ packet.sizes : num [1:4(1d)] 40401 40401 40401 40401
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ season: chr [1:4] "spring (MAM)" "summer (JJA)" "autumn (SON)" "winter (DJF)"
#> ..$ x.limits : num [1:2] -20.7 20.7
#> ..$ y.limits : num [1:2] -20.7 20.7
#> ..$ x.used.at : NULL
#> ..$ y.used.at : NULL
#> ..$ x.num.limit : NULL
#> ..$ y.num.limit : NULL
#> ..$ aspect.ratio : num 1
#> ..$ prepanel.default : chr "prepanel.default.levelplot"
#> ..$ prepanel : NULL
#> ..- attr(*, "class")= chr "trellis"
#> $ data: grouped_df[,4] [161,604 × 4] (S3: grouped_df/tbl_df/tbl/data.frame)
#> ..$ season: Ord.factor w/ 4 levels "spring (MAM)"<..: 1 1 1 1 1 1 1 1 1 1 ...
#> ..$ u : num [1:161604] -20.2 -20 -19.8 -19.6 -19.4 ...
#> ..$ v : num [1:161604] -20.2 -20.2 -20.2 -20.2 -20.2 ...
#> ..$ z : num [1:161604] NA NA NA NA NA NA NA NA NA NA ...
#> ..- attr(*, "groups")= tibble[,2] [4 × 2] (S3: tbl_df/tbl/data.frame)
#> .. ..$ season: Ord.factor w/ 4 levels "spring (MAM)"<..: 1 2 3 4
#> .. ..$ .rows : list<int> [1:4]
#> .. .. ..$ : int [1:40401] 1 2 3 4 5 6 7 8 9 10 ...
#> .. .. ..$ : int [1:40401] 40402 40403 40404 40405 40406 40407 40408 40409 40410 40411 ...
#> .. .. ..$ : int [1:40401] 80803 80804 80805 80806 80807 80808 80809 80810 80811 80812 ...
#> .. .. ..$ : int [1:40401] 121204 121205 121206 121207 121208 121209 121210 121211 121212 121213 ...
#> .. .. ..@ ptype: int(0)
#> .. ..- attr(*, ".drop")= logi TRUE
#> $ call: language polarPlot(mydata = openair::mydata, pollutant = "nox", type = "season")
#> - attr(*, "class")= chr "openair"
This produces the same side-by-side representation as you showed. Although it won't be obvious without knowing a lot more about how the graphical representation is handled, there does not appear to be any provision for the overplotting treatment that is being sought.
For a dataset as small as Rata_street_data, however, openair is overkill. I can try this in {ggplot2} if that's of interest.