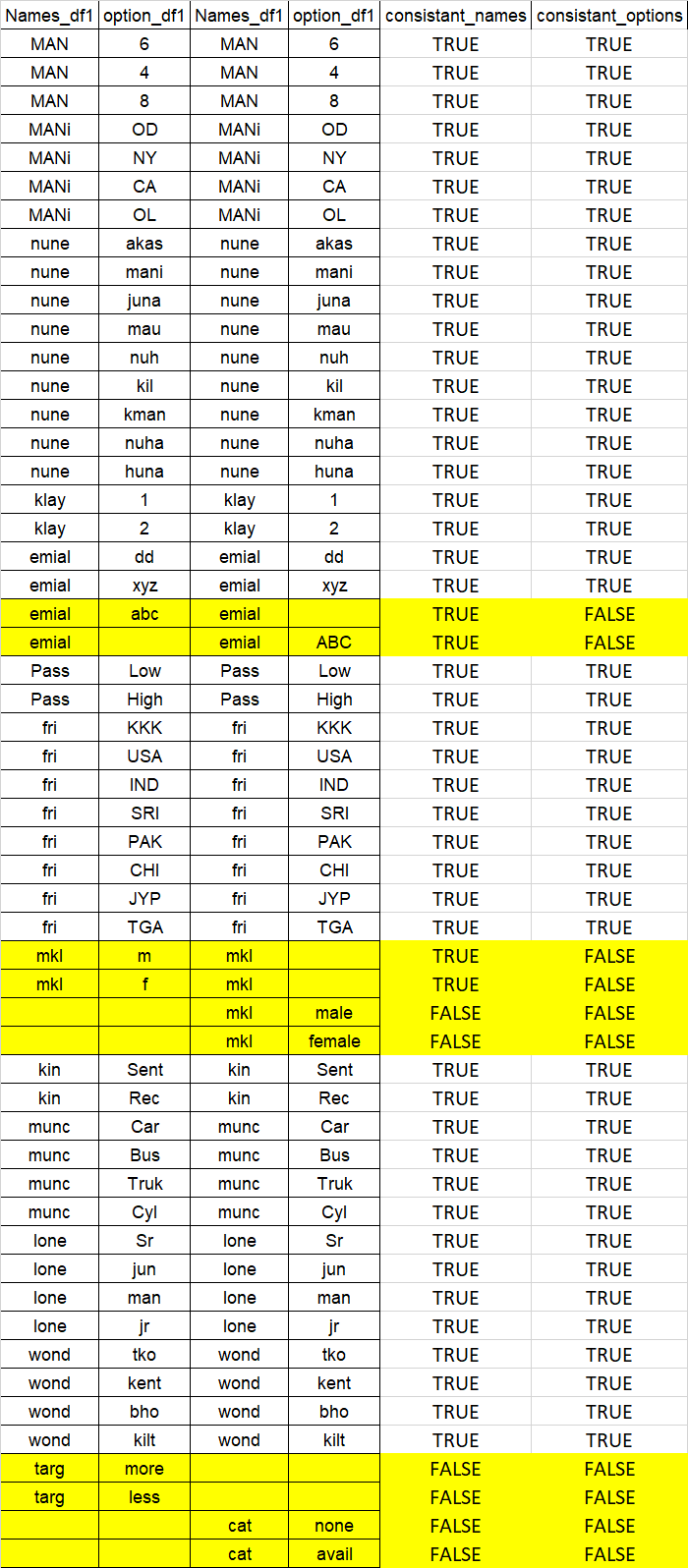
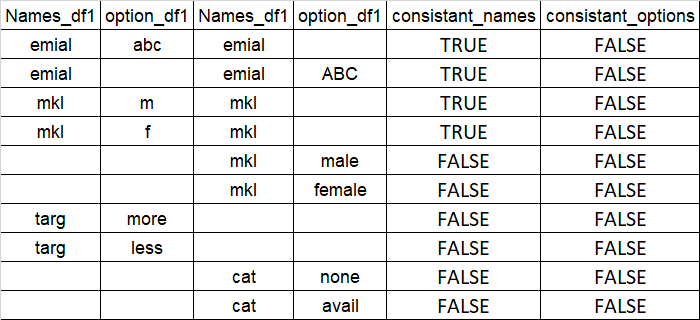
I have two data frames with same kind of data, now i want to check for all the columns in both data frames have same kind of text in all columns in both data frames .
so for example the column name "sales executives" in both data frames have exact name "Micheal klay" in both data frames but if there is any spelling error or extra space i want to show it as not matching.
I have tried below approach and its working for small database but because my data is very big, data having approx 10 - 40 millions or records so its showing error do we have any solution or any other approach to do that
cannot allocate vector of size 3.2GB
library(tidyverse)
df1 <- data.frame(MAN=c(6,6,4,6,8,6,8,4,4,6,6,8,8),MANi=c("OD","NY","CA","CA","OD","CA","OD","NY","OL","NY","OD","CA","OD"),
nune=c("akas","mani","juna","mau","nuh","kil","kman","nuha","huna","kman","nuha","huna","mani"),
klay=c(1,2,2,1,1,2,1,2,1,2,1,1,2),emial=c("dd","xyz","abc","dd","xyz","abc","dd","xyz","abc","dd","xyz","abc","dd"),Pass=c("Low","High","Low","Low","High","Low","High","High","Low","High","High","High","Low"),fri=c("KKK","USA","IND","SRI","PAK","CHI","JYP","TGA","KKK","USA","IND","SRI","PAK"),
mkl=c("m","f","m","m","f","m","m","f","m","m","f","m","m"),kin=c("Sent","Rec","Sent","Rec","Sent","Rec","Sent","Rec","Sent","Rec","Rec","Sent","Rec"),munc=c("Car","Bus","Truk","Cyl","Bus","Car","Bus","Bus","Bus","Car","Car","Cyl","Car"),
lone=c("Sr","jun","sr","jun","man","man","jr","Sr","jun","sr","jun","man","man"),wond=c("tko","kent","bho","kilt","kent","bho","kent","bho","bho","kilt","kent","bho","kilt"))
df2 <- data.frame(MAN=c(6,6,4,6,8,6,8,4,4,6,6,8,8,8,6),MANi=c("OD","NY","CA","CA","OD","CA","OD","NY","OL","ny","OD","CA","OD","NY","OL"),
nune=c("akas","mani","juna","mau","nuh","kil","kman","nuha","huna","kman","nuha","huna","mani","juna","mau"),
klay=c(1,2,2,1,1,2,1,2,1,2,1,1,2,2,1),emial=c("dd","xyz","ABC","dd","xyz","ABC","dd","xyz","ABC","dd","xyz","ABC","dd","xyz","ABC"),Pass=c("Low","High","Low","Low","High","Low","High","High","Low","High","High","High","Low","High","High"),fri=c("KKK","USA","IND","SRI","PAK","CHI","JYP","TGA","KKK","USA","IND","SRI","PAK","CHI","JYP"),
mkl=c("male","female","male","male","female","male","male","female","male","male","female","male","male","female","male"),kin=c("Sent","Rec","Sent","Rec","Sent","Rec","Sent","Rec","Sent","Rec","Rec","Sent","Rec","Sent","Rec"),munc=c("Car","Bus","Truk","Cyl","Bus","Car","Bus","Bus","Bus","Car","Car","Cyl","Car","Bus","Bus"),
lone=c("Sr","jun","sr","jun","man","man","jr","Sr","jun","sr","jun","man","man","jr","man"),wond=c("tko","kent","bho","kilt","kent","bho","kent","bho","bho","kilt","kent","bho","kilt","kent","bho"))
df1_long <- df1 %>%
as_tibble() %>%
mutate_if(is.double, as.character) %>% distinct() %>%
pivot_longer(everything(), names_to = "Names", values_to = "options") %>%
arrange(Names, options)
df2_long <- df2 %>%
as_tibble() %>%
mutate_if(is.double, as.character) %>% distinct() %>%
pivot_longer(everything(), names_to = "Names", values_to = "options") %>%
arrange(Names, options)
T1 <- df1_long %>%
full_join(df2_long, by=c("Names", "options"), keep = TRUE) %>%
distinct(Names.x, options.x, Names.y, options.y) %>%
arrange(Names.x, Names.y, options.x, options.y) %>%
mutate(
consistant_names = !is.na(Names.x) & !is.na(Names.y),
consistant_options = !is.na(options.x) & !is.na(options.y)
)
the output required like below
below are inconsistency between data bases