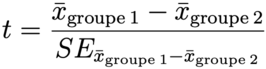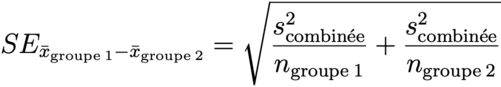d <- data.frame(
mpg = c(
22.8, 18.7, 14.3, 24.4, 22.8, 16.4, 17.3,
15.2, 10.4, 10.4, 14.7, 32.4, 30.4, 33.9,
21.5, 15.5, 15.2, 13.3,19.2, 27.3, 26,
30.4, 15.8, 15, 21.4
),
cyl = structure(c(
1,2, 2, 1, 1, 2, 2, 2, 2, 2, 2, 1, 1, 1,
1, 2, 2, 2, 2, 1, 1, 1, 2, 2, 1
), levels = c("4", "8"), class = "factor")
)
# classic t test
t.test(d$mpg ~ d$cyl, var.equal = TRUE)
#>
#> Two Sample t-test
#>
#> data: d$mpg by d$cyl
#> t = 8.1024, df = 23, p-value = 3.446e-08
#> alternative hypothesis: true difference in means between group 4 and group 8 is not equal to 0
#> 95 percent confidence interval:
#> 8.611261 14.516012
#> sample estimates:
#> mean in group 4 mean in group 8
#> 26.66364 15.10000
# however equal variance assumption is violated
var.test(d$mpg~d$cyl)
#>
#> F test to compare two variances
#>
#> data: d$mpg by d$cyl
#> F = 3.1033, num df = 10, denom df = 13, p-value = 0.05925
#> alternative hypothesis: true ratio of variances is not equal to 1
#> 95 percent confidence interval:
#> 0.9549589 11.1197133
#> sample estimates:
#> ratio of variances
#> 3.103299
# use Welsh test; confidence interval is for difference
# and does not include 0
t.test(d$mpg ~ d$cyl)
#>
#> Welch Two Sample t-test
#>
#> data: d$mpg by d$cyl
#> t = 7.5967, df = 14.967, p-value = 1.641e-06
#> alternative hypothesis: true difference in means between group 4 and group 8 is not equal to 0
#> 95 percent confidence interval:
#> 8.318518 14.808755
#> sample estimates:
#> mean in group 4 mean in group 8
#> 26.66364 15.10000
# if asssumption that data are drawn from a normal distribution
# is doubtful, use the nonparametric test
wilcox.test(d$mpg ~ d$cyl)
#> Warning in wilcox.test.default(x = DATA[[1L]], y = DATA[[2L]], ...): cannot
#> compute exact p-value with ties
#>
#> Wilcoxon rank sum test with continuity correction
#>
#> data: d$mpg by d$cyl
#> W = 154, p-value = 2.775e-05
#> alternative hypothesis: true location shift is not equal to 0
Created on 2023-06-13 with reprex v2.0.2