Hi Nir
I´m trying to run a K medoids analysis given a dataset with the following structure.
df <- structure(list(Sexo = c(2, 1, 1, 2, 1, 2, 2, 1, 1, 2, 1, 1, 2,
2, 1, 2, 1, 2, 2, 2, 1, 2, 1, 2, 2, 2, 1, 1, 1, 1), Escolaridad = c(1,
3, 1, 1, 2, 2, 3, 7, 3, 3, 1, 4, 2, 2, 7, 1, 2, 7, 2, 2, 2, 4,
2, 3, 1, 3, 2, 1, 1, 1), edad_intervalo = c(4, 1, 6, 6, 4, 4,
2, 2, 4, 1, 1, 3, 3, 4, 6, 4, 4, 6, 2, 4, 5, 3, 5, 6, 6, 2, 6,
6, 5, 5), seguro_votar = c(1, 1, 1, 1, 2, 1, 2, 1, 1, 1, 1, 1,
1, 2, 2, 2, 1, 1, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1), principal_eleme = c(1,
1, 3, 1, 5, 1, 1, 2, 1, 4, 1, 1, 1, 1, 88, 3, 1, 3, 1, 1, 1,
1, 1, 1, 1, 3, 1, 1, 1, 2), partido_votaste = c(88, 5, 2, 88,
7, 1, 66, 5, 7, 7, 6, 3, 2, 66, 1, 1, 7, 4, 88, 2, 7, 2, 7, 88,
1, 5, 88, 2, 7, 2), votarias_mismo = c(2, 2, 2, 88, 1, 88, 2,
88, 1, 1, 2, 2, 1, 2, 1, 2, 2, 1, 88, 1, 1, 1, 1, 1, 1, 1, 88,
2, 1, 2), arrepientes = c(1, 2, 1, 2, 2, 2, 88, 88, 2, 2, 2,
1, 2, 2, 2, 2, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 88, 1, 2, 2), dispuesto_cambiar = c(1,
2, 1, 4, 4, 3, 2, 3, 4, 3, 1, 1, 4, 3, 2, 1, 1, 2, 2, 3, 4, 4,
3, 88, 2, 4, 1, 1, 4, 1), voto_partido = c(5, 8, 7, 77, 77, 77,
77, 77, 7, 7, 6, 4, 2, 2, 2, 5, 1, 4, 7, 2, 7, 2, 7, 77, 88,
5, 77, 7, 5, 5), voto_partido_segundo = c(1, 4, 7, 88, 7, 88,
88, 77, 7, 7, 4, 99, 7, 3, 4, 5, 2, 4, 5, 2, 7, 2, 7, 77, 88,
99, 77, 7, 1, 5), voto_partido_nunca = c(99, 2, 2, 88, 2, 88,
99, 99, 7, 3, 1, 11, 1, 1, 2, 2, 7, 1, 88, 4, 7, 5, 1, 2, 88,
5, 88, 2, 2, 2)), row.names = c(NA, -30L), class = c("tbl_df",
"tbl", "data.frame"))
df <- na.omit(df)
install.packages("factoextra")
#> Installing package into 'C:/Users/Carlos Javier/AppData/Local/R/win-library/4.2'
#> (as 'lib' is unspecified)
#> package 'factoextra' successfully unpacked and MD5 sums checked
#>
#> The downloaded binary packages are in
#> C:\Users\Carlos Javier\AppData\Local\Temp\RtmpkvQCpy\downloaded_packages
install.packages("NbClust")
#> Installing package into 'C:/Users/Carlos Javier/AppData/Local/R/win-library/4.2'
#> (as 'lib' is unspecified)
#> package 'NbClust' successfully unpacked and MD5 sums checked
#>
#> The downloaded binary packages are in
#> C:\Users\Carlos Javier\AppData\Local\Temp\RtmpkvQCpy\downloaded_packages
library(factoextra)
#> Loading required package: ggplot2
#> Welcome! Want to learn more? See two factoextra-related books at https://goo.gl/ve3WBa
library(NbClust)
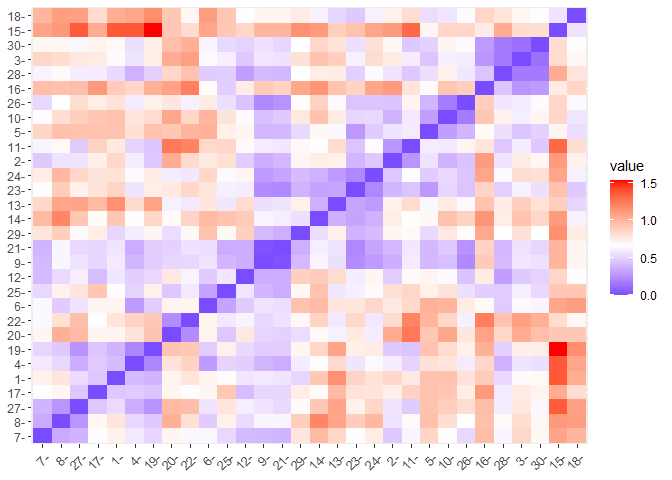
### Dissimilarity matrix between observations
m.distance <- get_dist(df, method = "kendall")
fviz_dist(m.distance, gradient = list(low = "blue", mid = "white", high = "red"))
### Optimal number of clusters
resnumclust<-NbClust(df, distance = "euclidean", min.nc=2, max.nc=10, method = "kmean", index = "all")
#> Warning in pf(beale, pp, df2): NaNs produced
#> Warning in pf(beale, pp, df2): NaNs produced
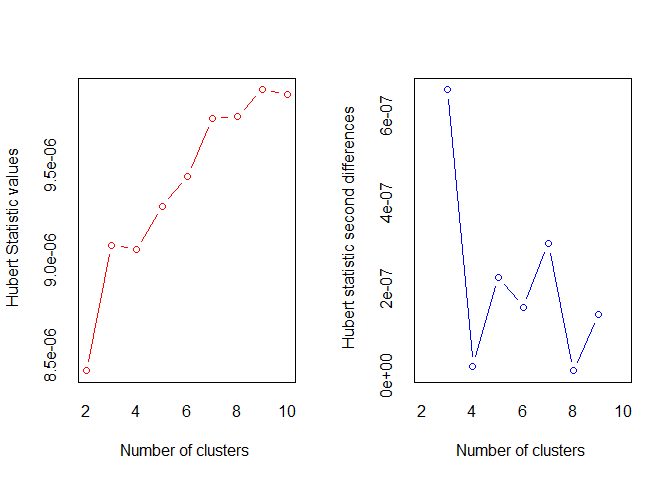
#> *** : The Hubert index is a graphical method of determining the number of clusters.
#> In the plot of Hubert index, we seek a significant knee that corresponds to a
#> significant increase of the value of the measure i.e the significant peak in Hubert
#> index second differences plot.
#>
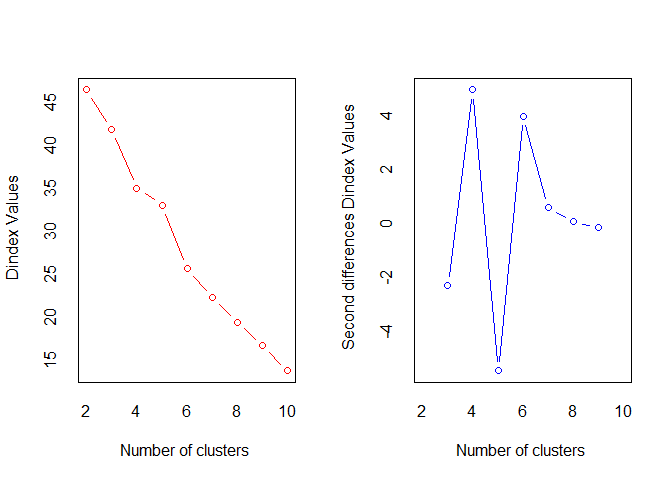
#> *** : The D index is a graphical method of determining the number of clusters.
#> In the plot of D index, we seek a significant knee (the significant peak in Dindex
#> second differences plot) that corresponds to a significant increase of the value of
#> the measure.
#>
#> *******************************************************************
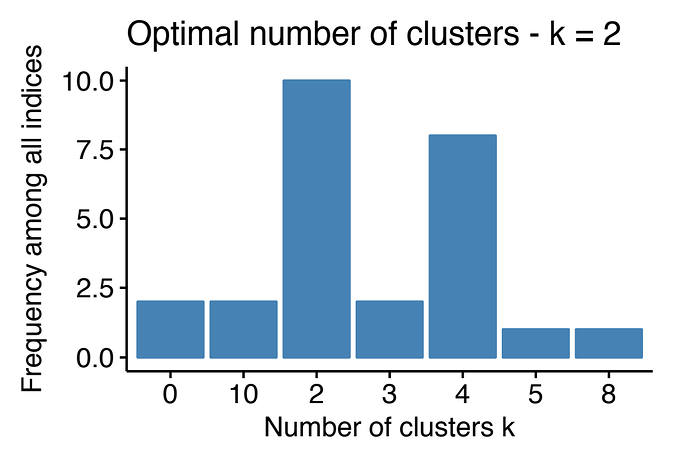
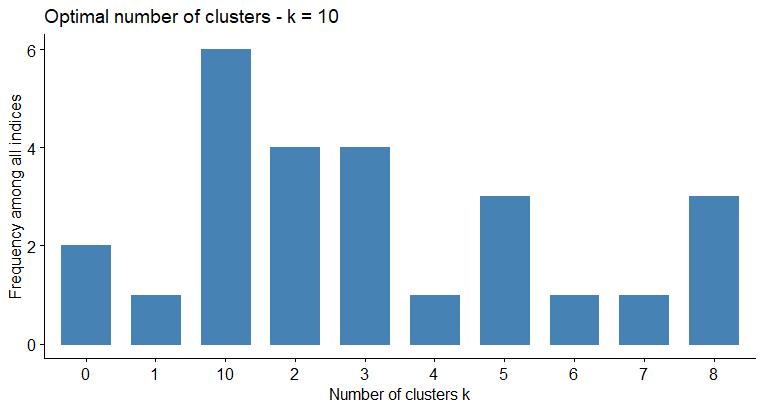
#> * Among all indices:
#> * 4 proposed 2 as the best number of clusters
#> * 4 proposed 3 as the best number of clusters
#> * 1 proposed 4 as the best number of clusters
#> * 3 proposed 5 as the best number of clusters
#> * 1 proposed 6 as the best number of clusters
#> * 1 proposed 7 as the best number of clusters
#> * 3 proposed 8 as the best number of clusters
#> * 6 proposed 10 as the best number of clusters
#>
#> ***** Conclusion *****
#>
#> * According to the majority rule, the best number of clusters is 10
#>
#>
#> *******************************************************************
resnumclust
#> $All.index
#> KL CH Hartigan CCC Scott Marriot TrCovW TraceW
#> 2 4.7554 28.0623 7.8695 2.1659 273.7554 8.000515e+35 200889646 110277.21
#> 3 0.9202 21.1270 9.0589 1.9050 338.9060 2.051864e+35 119265356 86083.10
#> 4 2.4259 21.0213 4.4068 2.9920 495.6268 1.964537e+33 72543834 64456.94
#> 5 0.4664 18.7884 10.3931 3.0214 789.7206 1.696823e+29 58905410 55115.26
#> 6 2.2940 22.4237 5.4113 5.3914 933.2714 2.041217e+27 35930171 38930.77
#> 7 1.0612 22.8098 5.7918 6.4042 1136.1672 3.210439e+24 31306337 31768.06
#> 8 1.2593 24.2019 5.2974 7.7747 1170.6220 1.329732e+24 19617010 25377.56
#> 9 1.2745 25.7135 4.7495 7.8881 1331.1268 7.989596e+21 17522959 20452.75
#> 10 0.6688 27.1938 9.2904 8.7344 1393.9771 1.213914e+21 13160121 16680.25
#> Friedman Rubin Cindex DB Silhouette Duda Pseudot2 Beale
#> 2 359.4712 2.8484 0.2723 0.9989 0.5747 0.6895 3.1516 3.2169
#> 3 401.6178 3.6490 0.2519 1.3410 0.5635 0.9151 1.8559 0.7199
#> 4 1067.9562 4.8733 0.2292 1.0237 0.5779 13.1637 -18.4807 0.0000
#> 5 24698.5989 5.6992 0.3219 0.8715 0.5875 0.4996 19.0269 7.7685
#> 6 25467.4797 8.0686 0.2394 0.7927 0.6160 2.5824 -10.4169 -2.5018
#> 7 27758.6616 9.8878 0.1855 0.7195 0.6540 2.0732 -0.5177 -2.1136
#> 8 28218.2422 12.3777 0.2065 0.5935 0.6703 2.3813 -9.2809 -4.4580
#> 9 32882.1535 15.3581 0.1485 0.5582 0.6984 8.3175 0.0000 0.0000
#> 10 35272.4036 18.8315 0.1438 0.4993 0.7443 2.6739 -0.6260 -2.5559
#> Ratkowsky Ball Ptbiserial Frey McClain Dunn Hubert SDindex Dindex
#> 2 0.3005 55138.603 0.7958 0.0929 0.2537 0.5122 0 0.0313 46.4604
#> 3 0.2927 28694.365 0.8548 0.9231 0.2567 0.5580 0 0.0352 41.8810
#> 4 0.2790 16114.235 0.8548 1.2299 0.2982 0.6101 0 0.0314 35.0033
#> 5 0.2956 11023.052 0.8728 0.7938 0.2616 0.6713 0 0.0277 33.0902
#> 6 0.2820 6488.462 0.8766 0.6309 0.2837 0.7023 0 0.0252 25.7111
#> 7 0.3015 4538.294 0.8772 0.3033 0.2715 0.6854 0 0.0258 22.3147
#> 8 0.2860 3172.194 0.8799 0.7126 0.2655 0.7981 0 0.0241 19.5014
#> 9 0.2757 2272.528 0.8687 0.3908 0.2378 0.6534 0 0.0274 16.7521
#> 10 0.2661 1668.025 0.8692 0.2617 0.2334 0.6534 0 0.0249 13.8567
#> SDbw
#> 2 0.7439
#> 3 0.8326
#> 4 0.6728
#> 5 0.4923
#> 6 0.3883
#> 7 0.3253
#> 8 0.2638
#> 9 0.2296
#> 10 0.1432
#>
#> $All.CriticalValues
#> CritValue_Duda CritValue_PseudoT2 Fvalue_Beale
#> 2 0.5264 6.2967 0.0008
#> 3 0.6717 9.7767 0.7314
#> 4 0.0549 344.0853 NaN
#> 5 0.6717 9.2879 0.0000
#> 6 0.2186 60.7604 1.0000
#> 7 0.2186 3.5741 1.0000
#> 8 0.6496 8.6302 1.0000
#> 9 0.0549 0.0000 NaN
#> 10 0.2186 3.5741 1.0000
#>
#> $Best.nc
#> KL CH Hartigan CCC Scott Marriot TrCovW
#> Number_clusters 2.0000 2.0000 5.0000 10.0000 5.0000 3.000000e+00 3
#> Value_Index 4.7554 28.0623 5.9863 8.7344 294.0938 3.916432e+35 81624291
#> TraceW Friedman Rubin Cindex DB Silhouette Duda
#> Number_clusters 4.00 5.00 6.0000 10.0000 10.0000 10.0000 2.0000
#> Value_Index 12284.48 23630.64 -0.5501 0.1438 0.4993 0.7443 0.6895
#> PseudoT2 Beale Ratkowsky Ball PtBiserial Frey McClain
#> Number_clusters 2.0000 3.0000 7.0000 3.00 8.0000 1 10.0000
#> Value_Index 3.1516 0.7199 0.3015 26444.24 0.8799 NA 0.2334
#> Dunn Hubert SDindex Dindex SDbw
#> Number_clusters 8.0000 0 8.0000 0 10.0000
#> Value_Index 0.7981 0 0.0241 0 0.1432
#>
#> $Best.partition
#> [1] 4 7 7 2 5 2 3 3 7 7 7 6 7 7 9 7 7 7 10 7 7 7 7 8 1
#> [26] 6 3 7 7 7
fviz_nbclust(resnumclust)
#> Error in if (class(best_nc) == "numeric") print(best_nc) else if (class(best_nc) == : the condition has length > 1
Created on 2022-08-14 by the reprex package (v2.0.1.9000)
The last line is the error when I try to visualize the optimal number of clusters (it should looks like in the image below)
Thank you very much