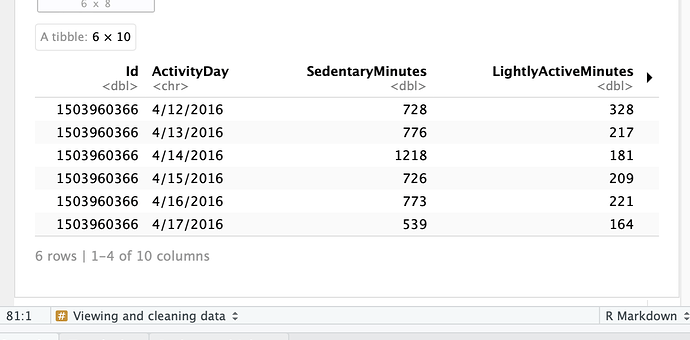
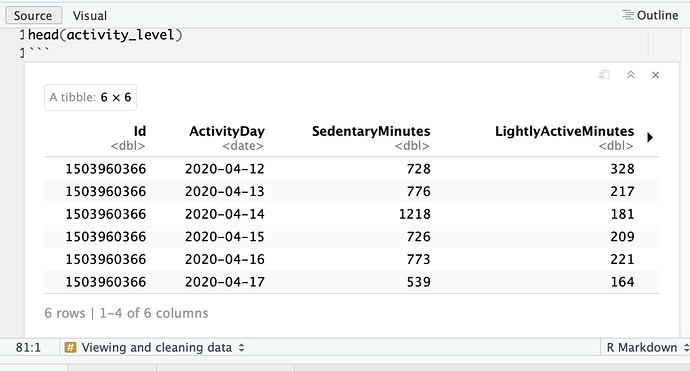
I get this:
Rows: 940 Columns: 10── Column specification ─────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (1): ActivityDay
dbl (9): Id, SedentaryMinutes, LightlyActiveMinutes, FairlyActiveMinutes, VeryActiveMinutes, SedentaryActiveD...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.structure(list(Id = c(1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366), ActivityDay = c("4/12/2016", "4/13/2016",
"4/14/2016", "4/15/2016", "4/16/2016", "4/17/2016"), SedentaryMinutes = c(728,
776, 1218, 726, 773, 539), LightlyActiveMinutes = c(328, 217,
181, 209, 221, 164), FairlyActiveMinutes = c(13, 19, 11, 34,
10, 20), VeryActiveMinutes = c(25, 21, 30, 29, 36, 38), SedentaryActiveDistance = c(0,
0, 0, 0, 0, 0), LightActiveDistance = c(6.05999994277954, 4.71000003814697,
3.91000008583069, 2.82999992370605, 5.03999996185303, 2.50999999046326
), ModeratelyActiveDistance = c(0.550000011920929, 0.689999997615814,
0.400000005960464, 1.25999999046326, 0.409999996423721, 0.779999971389771
), VeryActiveDistance = c(1.87999999523163, 1.57000005245209,
2.44000005722046, 2.14000010490417, 2.71000003814697, 3.19000005722046
)), row.names = c(NA, -6L), class = c("tbl_df", "tbl", "data.frame"
))