@williaml Here is a full code:
library(ggplot2)
library(reprex)
library(tidyverse)
library(reshape2)
#>
#> Attaching package: 'reshape2'
#> The following object is masked from 'package:tidyr':
#>
#> smiths
library(data.table)
#>
#> Attaching package: 'data.table'
#> The following objects are masked from 'package:reshape2':
#>
#> dcast, melt
#> The following objects are masked from 'package:dplyr':
#>
#> between, first, last
#> The following object is masked from 'package:purrr':
#>
#> transpose
library(scales)
#>
#> Attaching package: 'scales'
#> The following object is masked from 'package:purrr':
#>
#> discard
#> The following object is masked from 'package:readr':
#>
#> col_factor
#Help from R-community
df <- read.csv("F:/open_end_frequency_response.csv")
dt<- structure(list(modality = structure(c(1L, 2L, 3L, 5L, 4L), .Label = c("Appearance",
"Aroma", "Flavor", "Abstract", "Texture"), class = "factor"),
First_response = c(201L, 8L, 107L, 151L, 282L),
Second_response = c(72L, 17L, 148L, 225L, 260L),
Third_response = c(54L, 17L, 177L, 220L, 360L),
Fourth_response = c(46L, 24L, 168L, 198L, 356L ),
Fifth_response = c(39L, 13L, 122L, 150L, 402L),
Frequency = c(749L, 722L, 828L, 792L, 726L)), class = "data.frame",
row.names = c(NA,-5L))
dt1<-data.table(dt)[,.SD/Frequency,by=c("Frequency","modality"),.SDcols=2:6]
dat <- melt(dt1,id.vars = c("Frequency","modality"),variable.name = "Response", value.name = "Percentage",variable.factor=TRUE)
dat[,ggplot(.SD,aes(x = Response, y = Percentage, fill = modality)) +
geom_col(position = "dodge") +
geom_text(aes(label = scales::percent(Percentage, accuracy = 0.1)),
position = position_dodge(width=0.9), vjust = -0.25) +
labs(x = NULL, y = "Percentage") +
scale_y_continuous(labels = scales::label_percent()) +
scale_fill_brewer('Variables', palette='Spectral') +
theme_bw() +
theme(legend.title = element_blank(),
legend.position = c(.3, .95),
legend.direction = "horizontal"),]
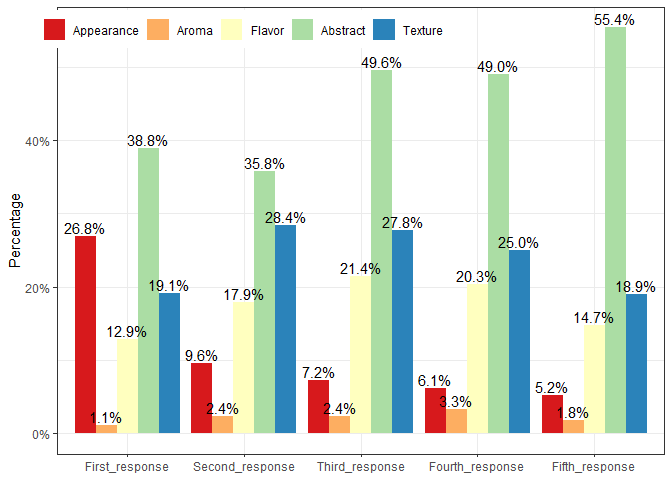
#another method
df <- read.csv("F:/open_end_frequency_response.csv")
colnames(df)[1] = "modality"
colnames(df)[2] = "First_response"
colnames(df)[3] = "Second_response"
colnames(df)[4] = "Third_response"
colnames(df)[5] = "Fourth_response"
colnames(df)[6] = "Fifth_response"
dput(df)
#> structure(list(modality = c("Appearance", "Aroma", "Flavor",
#> "Texture", "Abstract"), First_response = c(201L, 8L, 107L, 151L,
#> 282L), Second_response = c(72L, 17L, 148L, 225L, 260L), Third_response = c(54L,
#> 17L, 177L, 220L, 360L), Fourth_response = c(46L, 24L, 168L, 198L,
#> 356L), Fifth_response = c(39L, 13L, 122L, 150L, 402L), Frequency = c(412L,
#> 79L, 722L, 944L, 1660L)), class = "data.frame", row.names = c(NA,
#> -5L))
colnames(df) <- c("modality","First_response","Second_response","Third_response",
"Fourth_response", "Fifth_response", "Frequency")
ggplot(df, aes(x = modality))+
geom_bar()

#Help from R-community
df2 <- select(df, -Frequency)
dat <- melt(df2)
#> Warning in melt(df2): The melt generic in data.table has been passed a
#> data.frame and will attempt to redirect to the relevant reshape2 method;
#> please note that reshape2 is deprecated, and this redirection is now
#> deprecated as well. To continue using melt methods from reshape2 while both
#> libraries are attached, e.g. melt.list, you can prepend the namespace like
#> reshape2::melt(df2). In the next version, this warning will become an error.
#> Using modality as id variables
#Using modality as id variables
ggplot(dat, aes(modality, value, fill=interaction(variable))) +
geom_bar(stat='identity', position='dodge') +
ggtitle("A: General OEQ") +
theme_bw() + theme(axis.text.x = element_text(angle=90, hjust=1, size = 10, face = "bold")) +
scale_fill_brewer('Open-end', palette='RdPu') + theme(legend.position = c(.51, .80)) +
theme(legend.title=element_blank()) +
xlab("") + ylab("Frequency") + theme(panel.border = element_blank()) +
theme(axis.line = element_line(colour = "black")) +
theme(legend.key.size = unit(1.0, "cm"), legend.key.width = unit(0.5,"cm"),
legend.key.height = unit(0.5,"cm")) +
theme(legend.text = element_text(size = 7))
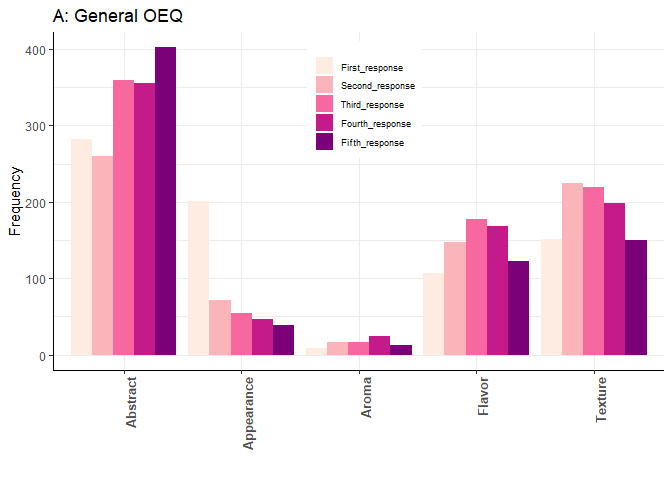
Created on 2021-02-05 by the reprex package (v0.3.0)


