Hello, andresrcs
install.packages('C:/Users/Sherwood/AppData/Local/Temp/RtmpkTfn52/downloaded_packages/devtools', repos = NULL, type="source")
Installing package into ‘C:/Users/Sherwood/Documents/R/win-library/3.6’
(as ‘lib’ is unspecified)
* installing *binary* package 'devtools' ...
cp: unknown option -- )
Try '/Rtools/bin/cp --help' for more information.
ERROR: installing binary package failed
* removing 'C:/Users/Sherwood/Documents/R/win-library/3.6/devtools'
* restoring previous 'C:/Users/Sherwood/Documents/R/win-library/3.6/devtools'
Warning in install.packages :
installation of package ‘C:/Users/Sherwood/AppData/Local/Temp/RtmpkTfn52/downloaded_packages/devtools’ had non-zero exit status
So it means it is not successful, isn't it?
Though it appears as a package in my computer
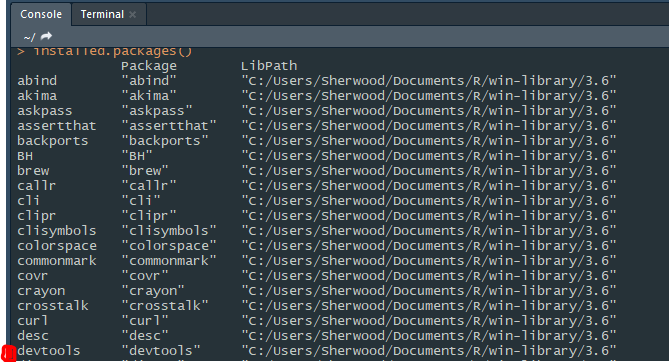 !
!
and show the imports:
devtools "callr, cli, covr (>= 3.2.0), crayon, desc, digest, DT,\nellipsis (>= 0.3.0), glue, git2r (>= 0.23.0), httr (>= 0.4),\njsonlite, memoise (>= 1.0.0), pkgbuild (>= 1.0.3), pkgload (>=\n1.0.2), rcmdcheck (>= 1.3.3), remotes (>= 2.1.0), rlang,\nroxygen2 (>= 6.1.1), rstudioapi (>= 0.7), rversions,\nsessioninfo (>= 1.1.1), stats, testthat (>= 2.1.1), tools,\nutils, withr"
and the suggests as well:
devtools "BiocManager, bitops, curl (>= 0.9), evaluate, foghorn (>=\n1.1.0), gmailr (> 0.7.0), knitr, lintr (>= 0.2.1), mockery,\npingr, MASS, pkgdown, Rcpp (>= 0.10.0), rhub (>= 1.0.2),\nrmarkdown, spelling (>= 1.1), whisker"
i try seeing in the library but prompts this message:
library(devtools)
Loading required package: usethis
Error: package or namespace load failed for ‘usethis’ in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]):
there is no package called ‘Rcpp’
Error: package ‘usethis’ could not be loaded
 !
!