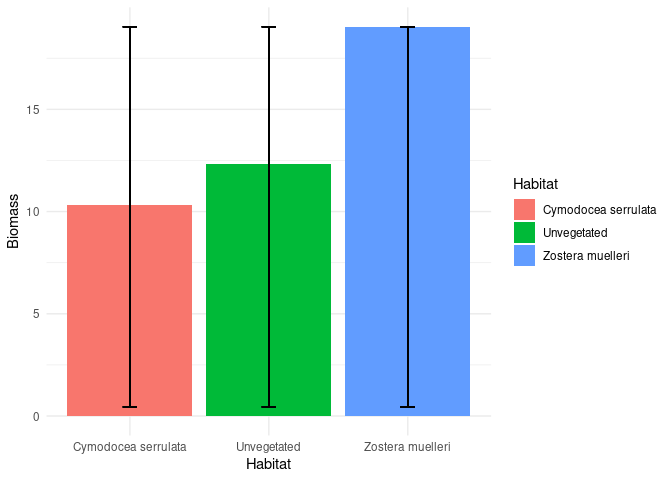
Here is @andresrcs code with these data
suppressPackageStartupMessages({
library(ggplot2)
})
dta <- data.frame(
Habitat = c("Cymodocea serrulata","Cymodocea serrulata",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Cymodocea serrulata",
"Unvegetated","Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Unvegetated","Unvegetated",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri","Unvegetated",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri","Unvegetated",
"Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Unvegetated","Unvegetated","Cymodocea serrulata",
"Cymodocea serrulata","Unvegetated",
"Cymodocea serrulata","Zostera muelleri","Unvegetated",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Cymodocea serrulata",
"Cymodocea serrulata","Unvegetated",
"Cymodocea serrulata","Cymodocea serrulata",
"Zostera muelleri","Zostera muelleri","Unvegetated",
"Unvegetated","Zostera muelleri",
"Zostera muelleri","Zostera muelleri","Unvegetated",
"Unvegetated","Unvegetated","Unvegetated",
"Unvegetated","Unvegetated",
"Zostera muelleri","Zostera muelleri","Zostera muelleri",
"Zostera muelleri","Zostera muelleri",
"Cymodocea serrulata","Zostera muelleri",
"Unvegetated","Zostera muelleri","Cymodocea serrulata",
"Cymodocea serrulata"),
Biomass = c(1.8110906572861,1.8110906572861,1.8110906572861,
1.8110906572861,1.8110906572861,1.8110906572861,
1.8110906572861,1.8110906572861,
1.8110906572861,1.8110906572861,3.5136858432153,
3.5136858432153,3.5136858432153,3.5136858432153,
3.5136858432153,3.5136858432153,
3.5136858432153,6.03853001889327,9.54472004811211,
14.1905516041284,14.1905516041284,14.1905516041284,
1.32397212016828,2.35125517813125,
0.657958442111442,1.34073024054044,1,1,1,1,1,1,
1,1,1,1,1,1,1,1.03850561425917,
1.03850561425917,1.03850561425917,2.05108844689479,
3.57673849471692,8.60102748523887,
12.3187184392026,0.912068005115458,0.912068005115458,
0.912068005115458,1.77741221332525,
1.77741221332525,3.06577363980793,7.24614312966712,
10.3051175546404,10.3051175546404,
10.3051175546404,1.8110906572861,1.8110906572861,
1.8110906572861,1.8110906572861,1.8110906572861,
1.8110906572861,3.5136858432153,
6.03853001889327,0.608519266879654,0.608519266879654,
0.608519266879654,0.608519266879654,
0.608519266879654,0.608519266879654,
0.608519266879654,0.608519266879654,0.608519266879654,
0.608519266879654,1.20992144244298,
1.20992144244298,1.20992144244298,2.12146275350218,
3.5136858432153,1,19.0398139780978,
19.0398139780978,0.472860390879922,0.453598006421229,
0.453598006421229,0.453598006421229,
0.905924859856634,0.905924859856634,0.444505251225668,
0.444505251225668,0.444505251225668,
0.852813632194663,1.45232352560218,1.45232352560218,
2.27796998852613,2.61254938723566,
1.02313525929414,2.10192512763994,1.8110906572861,
1.8110906572861)
)
p <- ggplot(dta,aes(Habitat,Biomass, fill = Habitat))
p + geom_bar(aes(Habitat,Biomass),position=position_dodge(.9), stat="identity") +
geom_errorbar(aes(ymin = min(Biomass), ymax = max(Biomass)), width = 0.1) +
xlab("Habitat") +
ylab("Biomass") + theme_minimal()