Take a look at the dtwclust package
library(dtwclust)
#> Loading required package: proxy
#>
#> Attaching package: 'proxy'
#> The following objects are masked from 'package:stats':
#>
#> as.dist, dist
#> The following object is masked from 'package:base':
#>
#> as.matrix
#> Loading required package: dtw
#> Loaded dtw v1.22-3. See ?dtw for help, citation("dtw") for use in publication.
#> dtwclust:
#> Setting random number generator to L'Ecuyer-CMRG (see RNGkind()).
#> To read the included vignettes type: browseVignettes("dtwclust").
#> See news(package = "dtwclust") after package updates.
data(uciCT)
# Fuzzy preprocessing: calculate autocorrelation up to 50th lag
acf_fun <- function(series, ...) {
lapply(series, function(x) {
as.numeric(acf(x, lag.max = 50, plot = FALSE)$acf)
})
}
# Define overall configuration
cfgs <- compare_clusterings_configs(
types = c("p", "h", "f", "t"),
k = 19L:20L,
controls = list(
partitional = partitional_control(
iter.max = 30L,
nrep = 1L
),
hierarchical = hierarchical_control(
method = "all"
),
fuzzy = fuzzy_control(
# notice the vector
fuzziness = c(2, 2.5),
iter.max = 30L
),
tadpole = tadpole_control(
# notice the vectors
dc = c(1.5, 2),
window.size = 19L:20L
)
),
preprocs = pdc_configs(
type = "preproc",
# shared
none = list(),
zscore = list(center = c(FALSE)),
# only for fuzzy
fuzzy = list(
acf_fun = list()
),
# only for tadpole
tadpole = list(
reinterpolate = list(new.length = 205L)
),
# specify which should consider the shared ones
share.config = c("p", "h")
),
distances = pdc_configs(
type = "distance",
sbd = list(),
fuzzy = list(
L2 = list()
),
share.config = c("p", "h")
),
centroids = pdc_configs(
type = "centroid",
partitional = list(
pam = list()
),
# special name 'default'
hierarchical = list(
default = list()
),
fuzzy = list(
fcmdd = list()
),
tadpole = list(
default = list(),
shape_extraction = list(znorm = TRUE)
)
)
)
# Number of configurations is returned as attribute
num_configs <- sapply(cfgs, attr, which = "num.configs")
cat(
"\nTotal number of configurations without considering optimizations:",
sum(num_configs),
"\n\n"
)
#>
#> Total number of configurations without considering optimizations: 56
# Define evaluation functions based on CVI: Variation of Information (only crisp partition)
vi_evaluators <- cvi_evaluators("VI", ground.truth = CharTrajLabels)
score_fun <- vi_evaluators$score
pick_fun <- vi_evaluators$pick
comparison_short <- compare_clusterings(CharTraj,
types = c("f"), configs = cfgs,
seed = 293L, trace = TRUE,
score.clus = score_fun, pick.clus = pick_fun,
return.objects = TRUE
)
#> =================================== Preprocessing series ===================================
#> -------------- Applying fuzzy preprocessings: --------------
#> preproc
#> 1 acf_fun
#> =================================== Performing fuzzy clusterings ===================================
#> -------------- Using configuration: --------------
#> k fuzziness iter.max delta symmetric version preproc distance centroid
#> 1 19, 20 2 30 0.001 FALSE 2 acf_fun L2 fcmdd
#>
#> Precomputing distance matrix...
#> Repetition 1 for k = 19
#> Iteration 1: Changes / Distsum = 100 / 45.56378
#> Iteration 2: Changes / Distsum = 0 / 45.56378
#> Repetition 1 for k = 20
#> Iteration 1: Changes / Distsum = 100 / 40.58559
#> Iteration 2: Changes / Distsum = 0 / 40.58559
#>
#> Elapsed time is 0.113 seconds.
#> -------------- Using configuration: --------------
#> k fuzziness iter.max delta symmetric version preproc distance centroid
#> 2 19, 20 2.5 30 0.001 FALSE 2 acf_fun L2 fcmdd
#>
#> Precomputing distance matrix...
#> Repetition 1 for k = 19
#> Iteration 1: Changes / Distsum = 100 / 41.39603
#> Iteration 2: Changes / Distsum = 0 / 41.39603
#> Repetition 1 for k = 20
#> Iteration 1: Changes / Distsum = 100 / 43.48574
#> Iteration 2: Changes / Distsum = 0 / 43.48574
#>
#> Elapsed time is 0.054 seconds.
require(doParallel)
#> Loading required package: doParallel
#> Loading required package: foreach
#> Loading required package: iterators
#> Loading required package: parallel
registerDoParallel(cl <- makeCluster(detectCores()))
comparison_long <- compare_clusterings(CharTraj,
types = c("p", "h", "f", "t"),
configs = cfgs,
seed = 293L, trace = TRUE,
score.clus = score_fun,
pick.clus = pick_fun,
return.objects = TRUE
)
#> =================================== Preprocessing series ===================================
#> -------------- Applying partitional preprocessings: --------------
#> preproc center_preproc
#> 1 none NA
#> 2 zscore FALSE
#> -------------- Applying hierarchical preprocessings: --------------
#> preproc center_preproc
#> 1 none NA
#> 2 zscore FALSE
#> -------------- Applying fuzzy preprocessings: --------------
#> preproc
#> 1 acf_fun
#> -------------- Applying tadpole preprocessings: --------------
#> preproc new.length_preproc
#> 1 reinterpolate 205
#> =================================== Performing partitional clusterings ===================================
#> =================================== Performing hierarchical clusterings ===================================
#> =================================== Performing fuzzy clusterings ===================================
#> =================================== Performing tadpole clusterings ===================================
# Using all external CVIs and majority vote
external_evaluators <- cvi_evaluators("external", ground.truth = CharTrajLabels)
score_external <- external_evaluators$score
pick_majority <- external_evaluators$pick
comparison_majority <- compare_clusterings(CharTraj,
types = c("p", "h", "f", "t"),
configs = cfgs,
seed = 84L, trace = TRUE,
score.clus = score_external,
pick.clus = pick_majority,
return.objects = TRUE
)
#> =================================== Preprocessing series ===================================
#> -------------- Applying partitional preprocessings: --------------
#> preproc center_preproc
#> 1 none NA
#> 2 zscore FALSE
#> -------------- Applying hierarchical preprocessings: --------------
#> preproc center_preproc
#> 1 none NA
#> 2 zscore FALSE
#> -------------- Applying fuzzy preprocessings: --------------
#> preproc
#> 1 acf_fun
#> -------------- Applying tadpole preprocessings: --------------
#> preproc new.length_preproc
#> 1 reinterpolate 205
#> =================================== Performing partitional clusterings ===================================
#> =================================== Performing hierarchical clusterings ===================================
#> =================================== Performing fuzzy clusterings ===================================
#> =================================== Performing tadpole clusterings ===================================
# best results
plot(comparison_majority$pick$object)
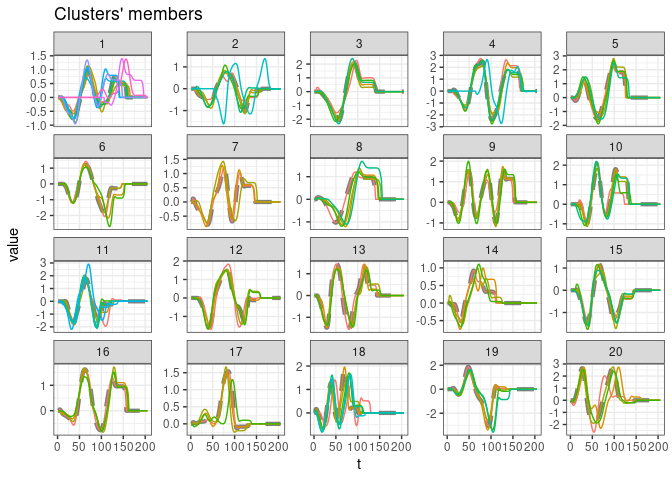
print(comparison_majority$pick$config)
#> config_id k dc window.size lb preproc new.length_preproc centroid
#> 4 config7_4 20 2 19 lbk reinterpolate 205 default
#> znorm_centroid ARI RI J FM VI
#> 4 NA 0.7574621 0.980202 0.6230769 0.768818 4.545371
stopCluster(cl)
registerDoSEQ()
# A run with only partitional clusterings
p_cfgs <- compare_clusterings_configs(
types = "p", k = 19L:21L,
controls = list(
partitional = partitional_control(
iter.max = 20L,
nrep = 8L
)
),
preprocs = pdc_configs(
"preproc",
none = list(),
zscore = list(center = c(FALSE, TRUE))
),
distances = pdc_configs(
"distance",
sbd = list(),
dtw_basic = list(
window.size = 19L:20L,
norm = c("L1", "L2")
),
gak = list(
window.size = 19L:20L,
sigma = 100
)
),
centroids = pdc_configs(
"centroid",
partitional = list(
pam = list(),
shape = list()
)
)
)
# Remove redundant (shape centroid always uses zscore preprocessing)
id_redundant <- p_cfgs$partitional$preproc == "none" &
p_cfgs$partitional$centroid == "shape"
p_cfgs$partitional <- p_cfgs$partitional[!id_redundant, ]
# LONG! 30 minutes or so, sequentially
# comparison_partitional <- compare_clusterings(CharTraj,
# types = "p",
# configs = p_cfgs,
# seed = 32903L, trace = TRUE,
# score.clus = score_fun,
# pick.clus = pick_fun,
# shuffle.configs = TRUE,
# return.objects = TRUE
# )
Created on 2021-01-06 by the reprex package (v0.3.0.9001)
