Hi community,
I have a dataset "dialong" which has the following variables-
id = individual id
time-id = index for time
age = age of individual
diab = if they have diabetic that year (0 or 1)
diabetes_data <- data.frame(
subjectid = c("a","b","c","d","e","f","g","h","i","j","k"),
age1 = c(27,18,28,14,11,32,20,19,12,23,34),
age2 = c(28,19,29,15,12,33,21,20,13,24,35),
age3 = c(29,20,30,16,13,34,22,21,14,25,36),
age4 = c(30,21,31,17,14,35,23,22,15,26,37),
age5 = c(31,22,32,18,15,36,24,23,16,27,38),
age6 = c(32,23,33,19,16,37,25,24,17,28,39),
age7 = c(33,24,34,20,17,38,26,25,18,29,40),
age8 = c(34,25,35,21,18,39,27,26,19,30,41),
age9 = c(35,26,36,22,19,40,28,27,20,31,42),
diab1 = c(0,0,0,0,0,0,0,0,0,0,0),
diab2 = c(0,1,0,0,0,0,0,0,0,0,0),
diab3 = c(0,NA,0,0,0,0,1,0,0,0,0),
diab4 = c(0,NA,0,0,0,0,NA,0,0,1,0),
diab5 = c(1,NA,0,0,0,0,NA,0,0,NA,0),
diab6 = c(NA,NA,0,0,0,0,NA,1,0,NA,0),
diab7 = c(NA,NA,0,0,1,0,NA,NA,0,NA,1),
diab8 = c(NA,NA,0,0,1,0,NA,NA,0,NA,NA),
diab9 = c(NA,NA,0,1,NA,0,NA,NA,0,NA,NA))
library(tidyr)
#> Warning: package 'tidyr' was built under R version 3.6.2
dialong <- pivot_longer(data = diabetes_data,cols = age1:diab9,
names_pattern = "([^\\d]+)(\\d+)",
names_to = c(".value","time_id"))
dialong <- drop_na(dialong)
Created on 2022-03-14 by the reprex package (v2.0.1)
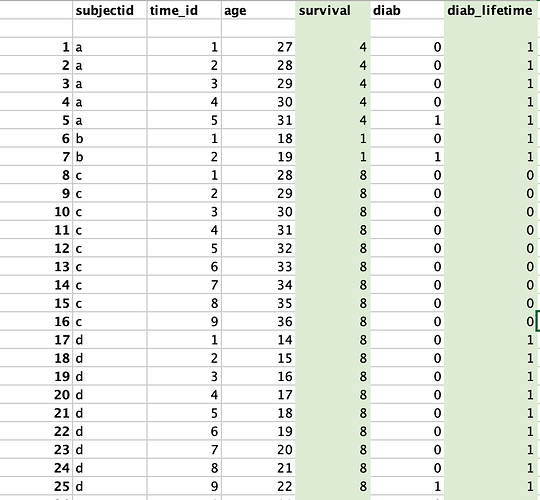
Now I want to create two new columns
-
survival time = age of entry - age of exit. This will be the same (non-time varying) throughout the data. e.g for subjectid "a", they enter in age 27 and leave at age 31. So, survival = 31 - 27 = 4.
-
diab_lifetime = the last value on "diab" condition for that individual. e.g. if indivdual has the value of diab = 1 in time-id "5" then their value of diab_lifetime = 1 through out the data. Note the individual is taken out the year they gets "1".
Here is how I want my final dataset to look (the new columns I want are highlighted in green)
Thank-you for the help !