Hi!
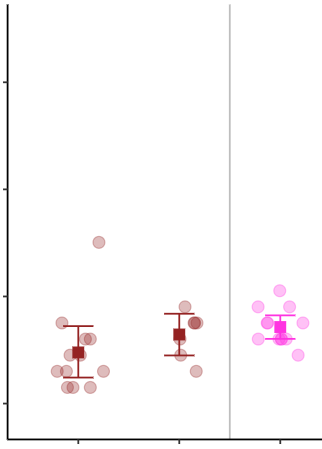
I updated my operating system and had to reinstall R, RStudio and all the packages I use. With the new version of this, when I render an html using Rmarkdown all my ggplots have a black background when I did not specify so. They look like this:
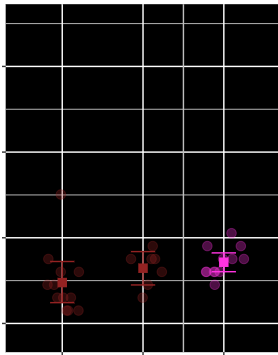
Here is the sessionInfo() of what I'm running:
sessionInfo()
R version 4.4.3 (2025-02-28 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8 LC_CTYPE=English_United States.utf8 LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C LC_TIME=English_United States.utf8
time zone: Europe/Berlin
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] here_1.0.1 brms_2.22.0 Rcpp_1.0.14 readr_2.1.5 tidyr_1.3.1 scales_1.4.0
[7] forcats_1.0.0 emmeans_1.11.0 dplyr_1.1.4 stringi_1.8.4 reactable_0.4.4 htmltools_0.5.8.1
[13] patchwork_1.3.0 ggbeeswarm_0.7.2 ggplot2_3.5.2 kableExtra_1.4.0
loaded via a namespace (and not attached):
[1] tensorA_0.36.2.1 gtable_0.3.6 beeswarm_0.4.0 xfun_0.51 htmlwidgets_1.6.4
[6] lattice_0.22-6 tzdb_0.5.0 vctrs_0.6.5 tools_4.4.3 generics_0.1.3
[11] parallel_4.4.3 sandwich_3.1-1 tibble_3.2.1 pkgconfig_2.0.3 Matrix_1.7-2
[16] checkmate_2.3.2 RColorBrewer_1.1-3 RcppParallel_5.1.10 distributional_0.5.0 lifecycle_1.0.4
[21] compiler_4.4.3 farver_2.1.2 stringr_1.5.1 Brobdingnag_1.2-9 codetools_0.2-20
[26] vipor_0.4.7 bayesplot_1.11.1 yaml_2.3.10 pillar_1.10.2 MASS_7.3-64
[31] bridgesampling_1.1-2 abind_1.4-8 multcomp_1.4-28 nlme_3.1-167 posterior_1.6.1
[36] tidyselect_1.2.1 digest_0.6.37 mvtnorm_1.3-3 purrr_1.0.4 splines_4.4.3
[41] rprojroot_2.0.4 fastmap_1.2.0 grid_4.4.3 cli_3.6.4 magrittr_2.0.3
[46] loo_2.8.0.9000 survival_3.8-3 TH.data_1.1-3 withr_3.0.2 backports_1.5.0
[51] estimability_1.5.1 rmarkdown_2.29 matrixStats_1.5.0 zoo_1.8-14 hms_1.1.3
[56] coda_0.19-4.1 evaluate_1.0.3 knitr_1.50 viridisLite_0.4.2 rstantools_2.4.0
[61] rlang_1.1.5 xtable_1.8-4 glue_1.8.0 xml2_1.3.8 svglite_2.1.3
[66] rstudioapi_0.17.1 R6_2.6.1 systemfonts_1.2.1