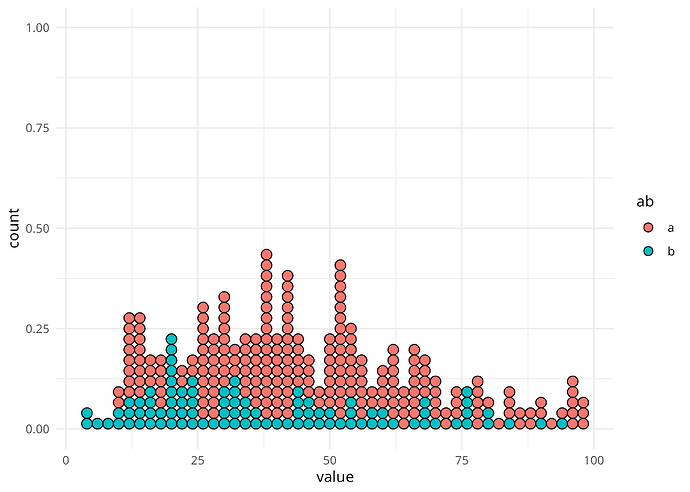
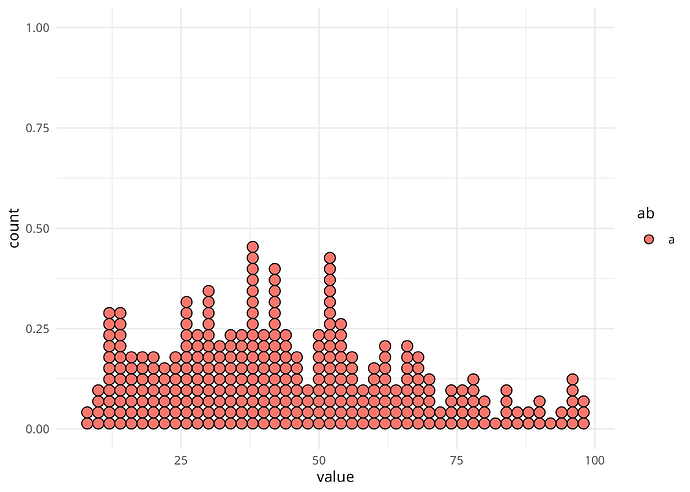
I am clearly misunderstanding something about how geom_dotplot works, or how the binwidth parameter should be understood, so I'd really welcome a clarification.
In brief: if I use ggplot2's geom_dotplot() (or similar functions from other packages, here ggdistfor reference), and set binwidth to 2, I'd expect to get some dots in every bin, unless there's a "hole" in my data, with no cases in a given bin of width 2.
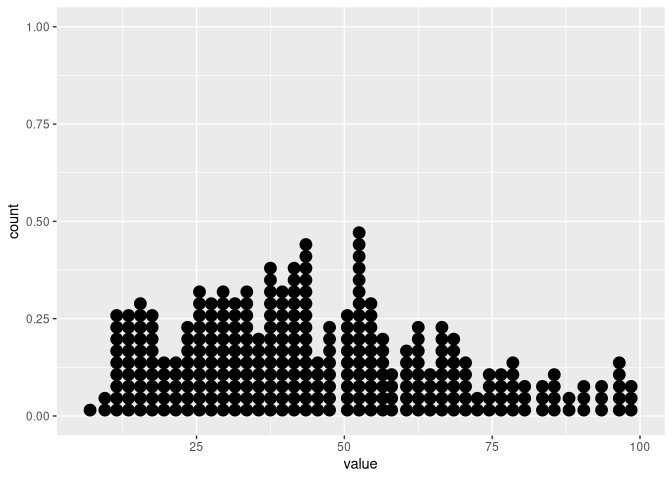
If you look at the graphs below, you will see that there is a visible "hole"/empty column just before value 50. However, in my data there is no bin of width 2 that is expected to be empty, as there are values for both 48 and 50.
I must be obviously misunderstanding something very basic about how binning (or one of the functions involved) works, but I couldn't figure out through the documentation what is the issue. I'd really welcome:
- any pointer to an explanation of how binning works in these cases, and why a bin with
binwidth2 should have value 0 if cutting data with width 2 does not not lead to any bin with value zero - a way to use
geom_dotplotthat reflects bins as they result fromcut
Thanks!
library("dplyr", warn.conflicts = FALSE)
library("ggplot2")
library("ggdist")
df <- data.frame(ab =c(rep("a", 308),
rep("b", 91)),
value =
c(56L, 14L, 11L, 12L, 37L, 17L, 61L, 19L, 79L, 81L, 67L, 13L,
15L, 28L, 62L, 54L, 45L, 29L, 15L, 54L, 27L, 17L, 13L, 62L, 44L,
88L, 31L, 96L, 42L, 21L, 28L, 52L, 94L, 52L, 34L, 66L, 32L, 99L,
54L, 37L, 57L, 20L, 15L, 39L, 65L, 27L, 52L, 58L, 18L, 33L, 52L,
69L, 64L, 24L, 42L, 23L, 54L, 38L, 71L, 69L, 43L, 56L, 47L, 16L,
62L, 55L, 48L, 41L, 12L, 76L, 18L, 45L, 51L, 38L, 29L, 63L, 58L,
15L, 18L, 12L, 42L, 11L, 74L, 98L, 99L, 66L, 39L, 21L, 34L, 42L,
55L, 38L, 25L, 46L, 23L, 28L, 78L, 53L, 19L, 21L, 41L, 24L, 12L,
52L, 68L, 43L, 50L, 39L, 84L, 32L, 23L, 24L, 32L, 21L, 55L, 62L,
23L, 47L, 34L, 9L, 48L, 41L, 44L, 27L, 15L, 74L, 20L, 26L, 68L,
44L, 34L, 47L, 43L, 41L, 67L, 64L, 97L, 26L, 51L, 31L, 66L, 43L,
61L, 96L, 17L, 37L, 71L, 46L, 53L, 26L, 18L, 31L, 35L, 17L, 14L,
79L, 48L, 35L, 66L, 63L, 52L, 40L, 33L, 20L, 28L, 39L, 44L, 53L,
16L, 15L, 38L, 50L, 14L, 47L, 41L, 57L, 61L, 38L, 30L, 84L, 60L,
44L, 75L, 22L, 67L, 31L, 29L, 90L, 54L, 23L, 13L, 51L, 46L, 50L,
26L, 68L, 51L, 40L, 43L, 85L, 83L, 86L, 30L, 34L, 38L, 96L, 42L,
36L, 74L, 77L, 44L, 62L, 39L, 38L, 43L, 25L, 36L, 31L, 71L, 53L,
86L, 65L, 33L, 97L, 39L, 30L, 25L, 60L, 80L, 40L, 81L, 85L, 57L,
30L, 26L, 73L, 12L, 51L, 57L, 25L, 36L, 13L, 48L, 79L, 31L, 58L,
15L, 30L, 27L, 57L, 52L, 34L, 13L, 14L, 16L, 33L, 69L, 76L, 42L,
88L, 41L, 53L, 42L, 60L, 70L, 72L, 52L, 38L, 11L, 93L, 54L, 39L,
36L, 76L, 12L, 91L, 36L, 30L, 37L, 53L, 26L, 43L, 32L, 70L, 55L,
94L, 69L, 63L, 67L, 39L, 18L, 27L, 43L, 53L, 78L, 44L, 38L, 30L,
7L, 29L, 26L, 28L, 51L, 10L, 52L, 34L, 90L, 58L, 60L, 84L, 24L,
44L, 68L, 28L, 14L, 55L, 30L, 26L, 12L, 21L, 16L, 22L, 22L, 11L,
16L, 14L, 69L, 33L, 25L, 6L, 55L, 34L, 30L, 59L, 15L, 13L, 16L,
22L, 38L, 20L, 20L, 8L, 18L, 34L, 16L, 33L, 25L, 21L, 21L, 25L,
30L, 21L, 5L, 37L, 33L, 63L, 81L, 76L, 76L, 37L, 30L, 48L, 33L,
11L, 94L, 45L, 52L, 21L, 42L, 50L, 19L, 40L, 58L, 55L, 33L, 24L,
23L, 12L, 45L, 46L, 20L, 68L, 77L, 21L, 48L, 75L, 47L, 61L, 51L,
56L, 34L, 71L, 90L, 45L, 67L, 3L, 76L, 46L, 81L))
df |>
ggplot(aes(x = value, fill = ab)) +
geom_dotplot(binwidth = 2, binpositions = "all")
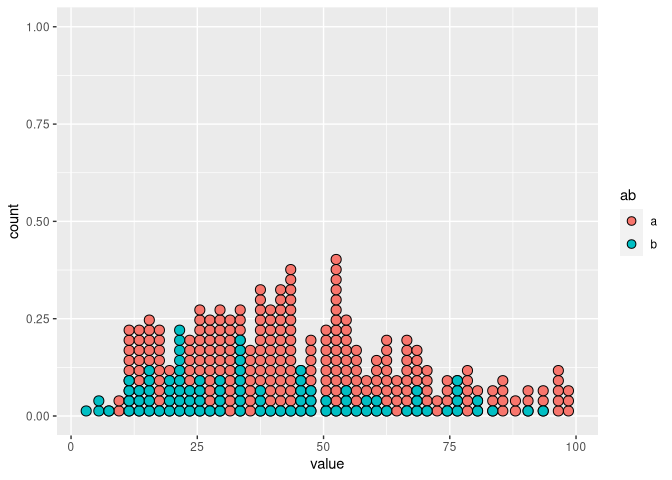
df |>
dplyr::filter(ab == "a") |>
ggplot(aes(x = value)) +
geom_dotplot(binwidth = 2, binpositions = "all")
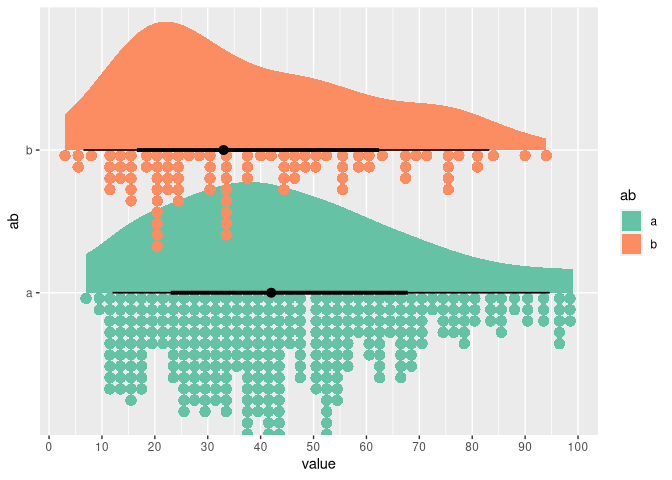
df %>%
ggplot(aes(y = ab, x = value, fill = ab)) +
stat_slab() +
stat_dotsinterval(aes(group = NA), side = "bottom", slab_linewidth = NA, binwidth = 2, position = position_dodge()) +
scale_x_continuous(n.breaks = 10) +
scale_fill_brewer(palette = "Set2")
df |>
dplyr::filter(ab == "a") |>
dplyr::group_by(value) |>
dplyr::count() |>
dplyr::filter(value>45)
#> # A tibble: 47 × 2
#> # Groups: value [47]
#> value n
#> <int> <int>
#> 1 46 3
#> 2 47 4
#> 3 48 4
#> 4 50 3
#> 5 51 6
#> 6 52 9
#> 7 53 7
#> 8 54 6
#> 9 55 4
#> 10 56 2
#> # ℹ 37 more rows
df |>
dplyr::filter(ab == "a") |>
dplyr::pull(value) |>
cut_width(width = 2, boundary = 0) |>
table()
#>
#> [6,8] (8,10] (10,12] (12,14] (14,16] (16,18] (18,20] (20,22]
#> 1 2 9 9 10 9 5 5
#> (22,24] (24,26] (26,28] (28,30] (30,32] (32,34] (34,36] (36,38]
#> 8 11 10 11 10 11 7 13
#> (38,40] (40,42] (42,44] (44,46] (46,48] (48,50] (50,52] (52,54]
#> 11 13 15 5 8 3 15 13
#> (54,56] (56,58] (58,60] (60,62] (62,64] (64,66] (66,68] (68,70]
#> 6 9 3 8 5 6 7 6
#> (70,72] (72,74] (74,76] (76,78] (78,80] (80,82] (82,84] (84,86]
#> 4 4 4 3 4 2 3 4
#> (86,88] (88,90] (90,92] (92,94] (94,96] (96,98] (98,100]
#> 2 2 1 3 3 3 2
Created on 2023-08-01 with reprex v2.0.2