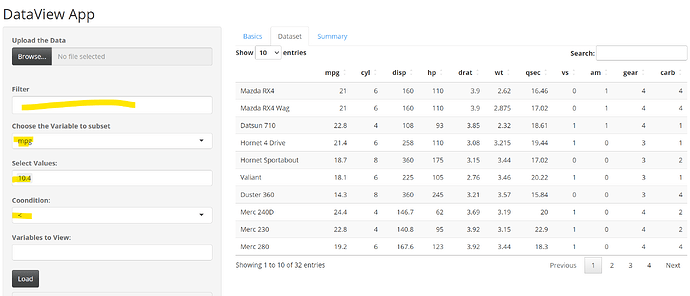
I am trying to create a simple app where i would like to auto populate the textinput based on the variable, values and condition selected from the dropdown
for suppose if i want to filter the data based on mpg < 10.4 then i would like to select them from the variable, values and condition option and then auto populate the mpg < 10.4 in the filter option and then when i click the load this filter should subset the data
also this filter should auto populate the values as i continue to select another condition like mpg <= 10.4 & cyl==8
could you please let me know your thoughts on the same, here is the code
library(shiny)
library(DT)
library(haven)
library(shinythemes)
library(DataExplorer)
library(labelled)
library(tidyverse)
# Define UI for application that draws a histogram
ui <- fluidPage(theme = shinytheme("spacelab"),
# Application title
titlePanel("DataView App"),
# Show a plot of the generated distribution
sidebarLayout(
sidebarPanel(
# Sidebar with a slider input for number of bins
fileInput("data",
"Upload the Data"),
textInput('tdata', 'Filter', value = NULL),
varSelectInput("variables1", "Choose the Variable to subset", NULL, selected = NULL, multiple = FALSE),
selectInput('var1','Select Values:',choices = '', selected = NULL, multiple = TRUE),
selectInput('cond1','Coondition:',choices = c('%in%','<','>','<=','>=','!=','&'), selected = NULL, multiple = FALSE),
varSelectInput("variables", "Variables to View:", NULL, multiple = TRUE),
actionButton("load","Load"),
h5(''),
verbatimTextOutput("input_dict")
), #sidebarPanel
mainPanel(
tabsetPanel(id='dataset',
tabPanel('Basics',verbatimTextOutput("input_intro")),
tabPanel('Dataset',DTOutput("input_file")),
tabPanel('Summary',verbatimTextOutput("input_sum"))
) #tabsetPanel
) #mainPanel
) #sidebarpanel
)
# Define server logic required to draw a histogram
server <- function(input, output, session) {
sampled <- reactive({
mtcars
})
observeEvent(input$variables1, {
updateSelectInput(session,inputId = "var1", 'Select Values:', choices = levels(as.factor(sampled()[[input$variables1]])),selected = NULL)
})
sampled2 <- reactive({
# req(input$variables1, input$var1)
if(input$variables1!='' & length(input$var1)>0){
sampled() %>% filter(CESEQ %in% input$var1)
} else if (input$variables1!='' & length(input$var1)==0){
sampled()
}
})
observe({
req(sampled())
updateVarSelectInput(session, "variables", "Variables to View:", sampled())
updateVarSelectInput(session, "variables1", "Choose the Variable to subset", sampled())
})
nsampled <- reactive({
# tdata2 <- unique(input$tdata)
# input$saveFilterButton
expre <- parse(text = input$tdata)
if(input$tdata != ''){
if (length(input$variables)==0) {
sampled() %>%
filter(eval(expre))
} else{
sampled() %>%
filter(eval(expre)) %>% dplyr::select(!!!input$variables)
}
} else {
if (length(input$variables)==0) {
sampled()
} else {
sampled() %>% dplyr::select(!!!input$variables)
}
}
}) %>%
bindEvent(input$load)
output$input_file <- renderDataTable({
# tdata2 <- unique(input$tdata)
# input$saveFilterButton
expre <- parse(text = input$tdata)
if(input$tdata != ''){
if (length(input$variables)==0) {
sampled() %>%
filter(eval(expre))
} else{
sampled() %>%
filter(eval(expre)) %>% dplyr::select(!!!input$variables)
}
} else {
if (length(input$variables)==0) {
sampled2()
} else {
sampled() %>% dplyr::select(!!!input$variables)
}
}
}) %>%
bindEvent(input$load)
output$input_sum <- renderPrint({
Hmisc::describe(nsampled())
}) %>%
bindEvent(input$load)
output$input_dict <- renderPrint({
labelled::generate_dictionary(nsampled()) %>% dplyr::select(variable, label, col_type)
}) %>%
bindEvent(input$load)
output$input_intro <- renderPrint({
nsampled2 <- DataExplorer::introduce(nsampled())
nsampled3 <- nsampled2 %>% pivot_longer(everything(), names_to = 'name3', values_to = 'Value') %>%
mutate(name2=stringr::str_to_sentence(name3), Name=stringr::str_replace_all(name2,'\\_',' ')) %>%
slice_head(n=5) %>% dplyr::select(Name, Value)
nsampled3
}) %>%
bindEvent(input$load)
}
# Run the application
shinyApp(ui = ui, server = server)