Hi everyone,
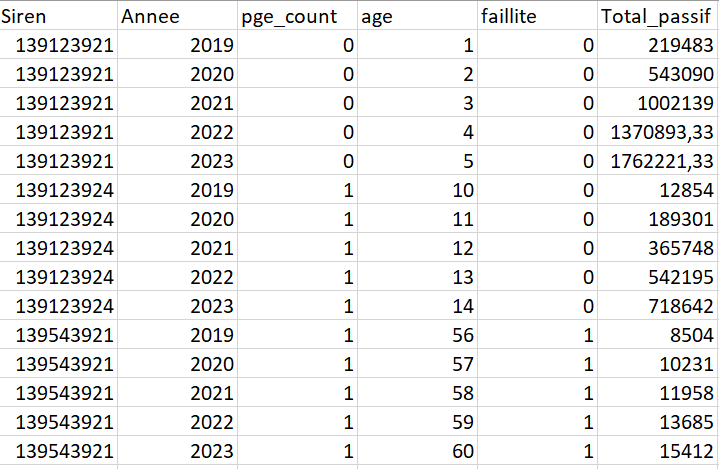
Here's my problem: I'm looking to analyse whether the granting of state-guaranteed loans during the crisis had an impact on the bankruptcy risk of the companies that took them out, and then also to analyse the impact of these loans on companies' classic financial variables. You will find the data structure in the appendix. To cut a long story short, when a company has gone bankrupt (default proceedings), it has a 1 in the bankruptcy column, 0 otherwise, when it has had at least one loan, it has a 1 in the pge_count column, and 0 otherwise.
My problem is that when I run the plm regression, I get errors every time, regardless of the model used or the indexes (Siren + Annee or Annee alone). Here's one such error: error in is.recursive(.$object) && !is.primitive(.$object) && n>0 : length = 2 in coercion to logical(1)
Otherwise, the matchit line is also empty. Is this due to the fact that I have 5 lines for each company and this creates a character that is too constant?
Here is my (short because i removed the lines to structure the data frame as a panel) code and you'll find the structure of the data at the end. Have in mind that i have approximately 3M lines.
Thank you for your help !
for (pge_var in pge_variables) {
wb <- createWorkbook()
# Génération des différentes possibilités de variables de contrôles
controle_combinations_faillite <- generate_controls_combinations(controle_vars_faillite)
controle_combinations <- generate_controls_combinations(controle_vars)
faillite_sheet <- "Faillite"
addWorksheet(wb, faillite_sheet)
iv_sheet <- "Faillite_IV_"
addWorksheet(wb, iv_sheet)
start_col <- 1
startIV_col <- 1
for (comb in controle_combinations_faillite) {
formule_faillite <- as.formula(
paste("faillite ~", pge_var, "+", paste(comb, collapse = " + "))
)
print(paste("Formule de faillite:", deparse(formule_faillite)))
# Convertir les données en pdata.frame
pdata <- pdata.frame(base_wide, index = c("Siren", "Annee"))
# Modèle à effets fixes
fe_model <- plm(formule_faillite, data = pdata, model = "within")
summary(fe_model)
# Modèle à effets aléatoires
re_model <- plm(formule_faillite, data = pdata, model = "random")
summary(re_model)
# Test de Hausman
hausman_test <- phtest(fe_model, re_model)
print(hausman_test)
# Régression IV
base_wide$prop_pge_secteur <- ave(base_wide[[pge_var]], base_wide$secteur, FUN = mean)
iv_formula <- as.formula(paste("faillite ~", pge_var, "+", paste(comb, collapse = " + "), "| prop_pge_secteur +", paste(comb, collapse = " + ")))
iv_model <- ivreg(iv_formula, data = base_wide)
# Conversion des résultats en data.frame
faillite_table <- as.data.frame(etable(fe_model))
writeData(wb, "Faillite", faillite_table, startCol = start_col)
start_col <- start_col + ncol(faillite_table) + 1
stargazer_file <- tempfile(fileext = ".txt")
stargazer(iv_model, type = "text", out = stargazer_file)
stargazer_content <- readLines(stargazer_file)
writeData(wb, iv_sheet, paste(stargazer_content, collapse = "\n"), startRow = 1, startCol = startIV_col)
startIV_col <- startIV_col + ncol(stargazer_content) + 1
}
# Régression 2 : Effet des PGE sur chaque variable d'intérêt
for (var in variables_interet) {
effet_sheet <- paste("Effet_", var, sep = "")
addWorksheet(wb, effet_sheet)
start_col <- 1
startIV_col <- 1
for (comb_pge in controle_combinations) {
formule_effet <- as.formula(
paste(var, "~", pge_var, "+", paste(comb_pge, collapse = " + "))
)
print(paste("Formule de variable d'intérêt : ", deparse(formule_effet)))
# Convertir les données en pdata.frame
pdata <- pdata.frame(base_wide, index = c("Siren", "Annee"))
# Modèle à effets fixes
fe_model <- plm(formule_effet, data = pdata, model = "within")
summary(fe_model)
# Modèle à effets aléatoires
re_model <- plm(formule_effet, data = pdata, model = "random")
summary(re_model)
# Test de Hausman
hausman_test <- phtest(fe_model, re_model)
print(hausman_test)
# Propensity Score Matching (PSM) pour essayer d'enlever le probable biais endogène
ps_model <- glm(as.formula(paste(pge_var, "~", paste(comb, collapse = " + "))),
data = base_wide, family = gaussian())
matched_data <- matchit(as.formula(paste(pge_var, "~", paste(comb, collapse = " + "))),
method = "nearest", data = base_wide)
matched_dataset <- match.data(matched_data)
# Régression sur le nouveau dataset apparié
psm_model <- lm(as.formula(paste(var, "~", pge_var, "+", paste(comb, collapse = " + "))),
data = matched_dataset)
effet_table <- as.data.frame(etable(fe_model))
psm_table <- as.data.frame(etable(psm_model))
effet_table <- cbind(effet_table, psm_table)
# Ajout sur feuille Excel
writeData(wb, effet_sheet, effet_table, startCol = start_col)
start_col <- start_col + ncol(effet_table) + 1
iv_sheet <- "Effet_IV_"
addWorksheet(wb, iv_sheet)
# Régression IV
iv_formula <- as.formula(paste("faillite ~", pge_var, "+", paste(comb, collapse = " + "), "| prop_pge_secteur +", paste(comb, collapse = " + ")))
iv_model <- ivreg(iv_formula, data = base_wide)
stargazer_file <- tempfile(fileext = ".txt")
stargazer(iv_model, type = "text", out = stargazer_file)
stargazer_content <- readLines(stargazer_file)
writeData(wb, iv_sheet, paste(stargazer_content, collapse = "\n"), startRow = 1, startCol = startIV_col)
}
}
output_file <- paste0(output_path, "Resultats_", pge_var, ".xlsx")
saveWorkbook(wb, output_file, overwrite = TRUE)
}