Is Dummy Score.csv a file that you made with your data? If so, it's not a good idea to store it inside the nparLD package directories. You should put your own files somewhere else (ideally, in a project folder — see: Project-oriented workflow).
I got the same error when trying the example from the JStatSoft article, but adding some NA values to the built-in dataset.
library("nparLD")
#> Loading required package: MASS
data("rat")
# Choose some rows where NAs will be added to `rat`
set.seed(42) # to make this code reproducible
NA_indices <- sample.int(length(rat$subject), size = 25)
# NAs for all variables
rat_NAall <- rat
rat_NAall[NA_indices, ] <- NA
nparLD(resp ~ time * group, data = rat_NAall, subject = "subject", description = FALSE)
#> Error in data.frame(dat, subject = subject): arguments imply differing number of rows: 110, 135
# NAs only for `resp` variable
rat_NAresp <- rat
rat_NAresp$resp[NA_indices] <- NA
nparLD(resp ~ time * group, data = rat_NAresp, subject = "subject", description = FALSE)
#> Error in data.frame(dat, subject = subject): arguments imply differing number of rows: 110, 135
I poked around in the nparLD() code a bit. Even though the documentation for the underlying model functions (such as f1.ld.f1()) says that the response variable can have NAs, nparLD() is written in a way that does not handle NA values gracefully. It could be altered to handle this situation better, but it would require changes in several different places. You might consider contacting the code maintainer (see CRAN for contact info) to ask if they'd be interested in making this change to the package — though the last update was 6 years ago, so it may not be an active project for them anymore.
It is possible to call the underlying model functions successfully when you have NAs in your response variable, but not when there are across-the-board NAs. The time, subject, and group variables need to have complete information (this makes sense, when you think about it).
f1.ld.f1(rat_NAall$resp, rat_NAall$time, rat_NAall$group, rat_NAall$subject)
#> Error in f1.ld.f1(rat_NAall$resp, rat_NAall$time, rat_NAall$group, rat_NAall$subject):
#> Number of levels of subject (28) times number of levels of time (6)
#> is not equal to the total number of observations (135).
f1.ld.f1(rat_NAresp$resp, rat_NAresp$time, rat_NAresp$group, rat_NAresp$subject)
#> Total number of observations: 110
#> Total number of subjects: 27
#> Total number of missing observations: 25
#>
#> Class level information
#> -----------------------
#> Levels of Time (sub-plot factor time) : 5
#> Levels of Group (whole-plot factor group) : 3
#>
#> Abbreviations
#> -----------------------
#> RankMeans = Rank means
#> Nobs = Number of observations
#> RTE = Relative treatment effect
#> case2x2 = tests for 2-by-2 design
#> Wald.test = Wald-type test statistic
#> ANOVA.test = ANOVA-type test statistic with Box approximation
#> ANOVA.test.mod.Box = modified ANOVA-type test statistic with Box approximation
#> Wald.test.time = Wald-type test statistic for simple time effect
#> ANOVA.test.time = ANOVA-type test statistic for simple time effect
#> N = Standard Normal Distribution N(0,1)
#> T = Student's T distribution with respective degrees of freedom
#> pattern.time (time effects) = Test against patterned alternatives in time using normal distribution ( no pattern specified )
#> pair.comparison = Tests for pairwise comparisions (without specifying a pattern)
#> pattern.pair.comparison = Test for pairwise comparisons with patterned alternatives in time ( no pattern specified )
#> pattern.group (group effects) = Test against patterned alternatives in group ( no pattern specified )
#> covariance = Covariance matrix
#> Note: The description output above will disappear by setting description=FALSE in the input. See the help file for details.
#>
#> F1 LD F1 Model
#> -----------------------
#> Check that the order of the time and group levels are correct.
#> Time level: 0 1 2 3 4
#> Group level: control thyrox thiour
#> If the order is not correct, specify the correct order in time.order or group.order.
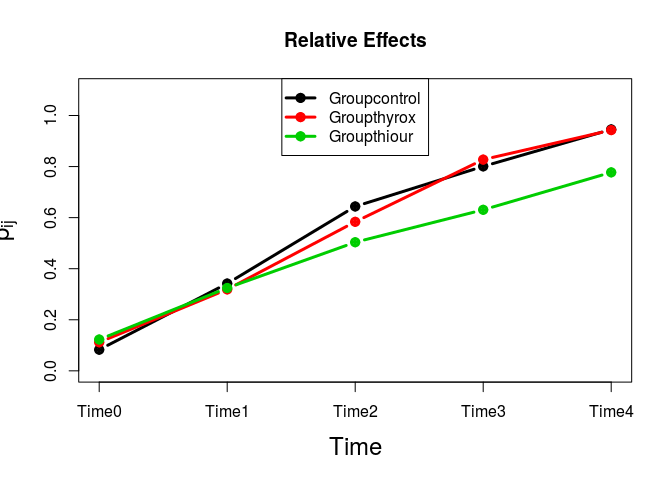
#> $RTE
#> RankMeans Nobs RTE
#> Groupcontrol 62.423611 37 0.56294192
#> Groupthyrox 61.756667 27 0.55687879
#> Groupthiour 52.395000 46 0.47177273
#> Time0 12.137037 23 0.10579125
#> Time1 36.631481 25 0.32846801
#> Time2 63.970833 23 0.57700758
#> Time3 83.319444 20 0.75290404
#> Time4 98.233333 19 0.88848485
#> Groupcontrol:Time0 9.611111 9 0.08282828
#> Groupcontrol:Time1 38.111111 9 0.34191919
#> Groupcontrol:Time2 71.312500 8 0.64375000
#> Groupcontrol:Time3 88.583333 6 0.80075758
#> Groupcontrol:Time4 104.500000 5 0.94545455
#> Groupthyrox:Time0 12.800000 5 0.11181818
#> Groupthyrox:Time1 35.583333 6 0.31893939
#> Groupthyrox:Time2 64.700000 5 0.58363636
#> Groupthyrox:Time3 91.500000 6 0.82727273
#> Groupthyrox:Time4 104.200000 5 0.94272727
#> Groupthiour:Time0 14.000000 9 0.12272727
#> Groupthiour:Time1 36.200000 10 0.32454545
#> Groupthiour:Time2 55.900000 10 0.50363636
#> Groupthiour:Time3 69.875000 8 0.63068182
#> Groupthiour:Time4 86.000000 9 0.77727273
#>
#> $case2x2
#> NULL
#>
#> $Wald.test
#> Statistic df p-value
#> Group 13.07227 2 1.450084e-03
#> Time 2666.63423 4 0.000000e+00
#> Group:Time 52.64966 8 1.260600e-08
#>
#> $ANOVA.test
#> Statistic df p-value
#> Group 4.565604 1.746226 0.013910038
#> Time 541.331813 2.874199 0.000000000
#> Group:Time 5.928531 4.751940 0.000026137
#>
#> $ANOVA.test.mod.Box
#> Statistic df1 df2 p-value
#> Group 4.565604 1.746226 14.9312 0.03226714
#>
#> $Wald.test.time
#> Statistic df p-value
#> control 2597.0149 4 0.000000e+00
#> thyrox 2710.8122 4 0.000000e+00
#> thiour 468.8299 4 3.687437e-100
#>
#> $ANOVA.test.time
#> Statistic df p-value
#> control 269.2295 2.306279 3.816710e-135
#> thyrox 162.5695 1.934844 4.138604e-69
#> thiour 137.4320 2.563663 1.485588e-76
#>
#> $pattern.time
#> NULL
#>
#> $pair.comparison
#> Pairs Test Statistic df p-value
#> 1 Groupcontrol:Groupthyrox Group 0.02724524 1.000000 8.688957e-01
#> 2 Groupcontrol:Groupthyrox Time 405.47440752 2.786149 6.604800e-245
#> 3 Groupcontrol:Groupthyrox Group:Time 1.15925983 2.786149 3.223463e-01
#> 4 Groupcontrol:Groupthiour Group 11.79919735 1.000000 5.925626e-04
#> 5 Groupcontrol:Groupthiour Time 393.19449475 2.760698 2.400405e-235
#> 6 Groupcontrol:Groupthiour Group:Time 9.96750698 2.760698 3.314825e-06
#> 7 Groupthyrox:Groupthiour Group 5.32069805 1.000000 2.107357e-02
#> 8 Groupthyrox:Groupthiour Time 297.80323245 2.442732 4.387837e-158
#> 9 Groupthyrox:Groupthiour Group:Time 7.49496054 2.442732 1.934505e-04
#>
#> $pattern.pair.comparison
#> NULL
#>
#> $covariance
#> NULL
#>
#> $model.name
#> [1] "F1 LD F1 Model"
Created on 2018-07-25 by the reprex package (v0.2.0).