Hi Posit Community,
I have been stuck on this issue with my R script for a significant time now, so I'm posting here hoping for some guidance on how to best manage this coding issue.
Some background first:
- I have main dataset with two columns that start with the name "term". From these two columns I'm trying to derive a new dataset by applying multiple text matching conditions with exclusions.
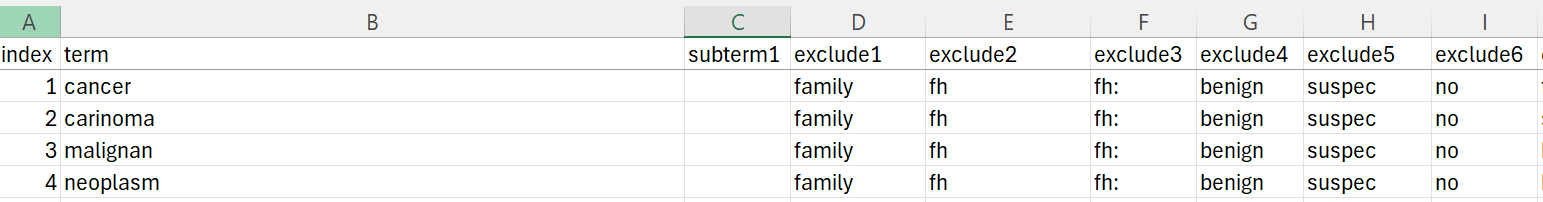
- I am using an excel sheet titled "searcterms_" with multiple columns including
- The term column, which contains the inclusion criteria for my inclusion (e.g., cancer) and multiple exclusion columns. Sometimes I'm utilising one inclusion term such as "cancer". Other times it's 2 or more words with spaces such as "cancer of the mouth"
- Subterm columns (subterm1, subterm2, etc.) - when sub terms are included alongside the corresponding main term, at least one of them MUST be present in the description for the code to be selected
- Exclusion columns (exclude1, exclude2, etc.) - if ANY of the exclusion terms are present, the code will not be selected
- This file also includes an index column (1, 2, 3, etc.)
NOTE: Search terms stored in table format because they are to be considered in some combination (i.e. an inclusion term with certain exclusion terms). I also want to include or exclude text that CONTAINS the characters of choice (e.g. exclude records that say "suspected", using the exclusion term "suspec".
***GOAL: include rows of the term variable in the "medical_combined" file if it contains any of the specified terms in the "searchterms_" file, however, exclude these terms if any of the exclusion terms are present, as specified in the exclude1 to excludeN columns in "searchterms_" file
***Problem with script: in the final output, records are being included despite containing the exclusion terms. I also want to make sure that all my inclusion terms (under columns term and subterm) are actually being considered for inclusion.
There may be issues with spaces, exact matching, partial matching, etc. I have explored multiple solutions, but none worked properly. I have previously edited the script and it became too restrictive (i.e. inclusion terms were not being included), or not restrictive enough (absolutely irrelevant records were being included).
Please help me resolve this issue, and make the R script run more efficiently and accurately where possible (sometimes I have hundreds of inclusion terms and corresponding exclusion terms).
I am providing my R script below (the part of the code where I think I'm running into problems). If you have any follow-up questions, please let me know Thank you so much for your time in advance! Much appreciated ![]()
EXAMPLE DATA:
-
searchterms_cancer file:
-
medical_combined file is an R file. I'm using character variables that start with the word "term"
# Step 3: Text Standardization (lowercase, remove extra spaces)
medical_combined <- medical_combined %>%
mutate(across(contains("term"), ~ tolower(str_trim(.))))
# Step 4: Import and structure search terms, exclusion terms
search_type <- "cancer" # Example search type, adjust as needed
search_terms <- read_excel(file.path(dir, paste0("searchterms_", search_type, ".xlsx")))
# Standardize text in search_terms
search_terms <- search_terms %>%
mutate(across(where(is.character), ~ tolower(str_trim(.))))
# Extract counts of search and exclusion terms
num_main_terms <- nrow(search_terms)
num_subterms <- sum(grepl("subterm", names(search_terms)))
num_exclusions <- sum(grepl("exclude", names(search_terms)))
# Step 5: Initialize candidate identification
# Initialize candidate column in medical_combined
medical_combined$candidate <- 0 # Set default to 0
# Loop through each main search term
for (i in seq_len(num_main_terms)) {
# Extract and prepare the main term
main_term <- tolower(str_trim(search_terms$term[i]))
# Subterm matching conditions
subterm_matches <- sapply(1:num_subterms, function(k) {
if (paste0("subterm", k) %in% names(search_terms)) {
as.numeric(grepl(main_term, medical_combined[[paste0("subterm", k)]], ignore.case = TRUE))
} else {
rep(0, nrow(medical_combined)) # Fill with 0 if subterm column is missing
}
})
# Exclusion matching conditions with boundary checks
exclusion_matches <- sapply(1:num_exclusions, function(j) {
exclusion_term <- tolower(str_trim(search_terms[[paste0("exclude", j)]]))
if (paste0("exclude", j) %in% names(search_terms)) {
as.numeric(grepl(paste0("\\b", exclusion_term, "\\b"), medical_combined[[paste0("exclude", j)]], ignore.case = TRUE))
} else {
rep(0, nrow(medical_combined)) # Fill with 0 if exclusion column is missing
}
})
# Ensure subterm_matches and exclusion_matches are data frames and have correct dimensions
subterm_matches_df <- as.data.frame(subterm_matches)
exclusion_matches_df <- as.data.frame(exclusion_matches)
# Handle empty data frames by setting the sums to zero if no columns are present
num_subterms_found <- if (ncol(subterm_matches_df) > 0) rowSums(subterm_matches_df, na.rm = TRUE) else rep(0, nrow(medical_combined))
num_exclusions_found <- if (ncol(exclusion_matches_df) > 0) rowSums(exclusion_matches_df, na.rm = TRUE) else rep(0, nrow(medical_combined))
# Ensure the result vectors have the same length as nrow(medical_combined)
if (length(num_subterms_found) != nrow(medical_combined)) {
num_subterms_found <- rep(0, nrow(medical_combined))
}
if (length(num_exclusions_found) != nrow(medical_combined)) {
num_exclusions_found <- rep(0, nrow(medical_combined))
}
# Debug outputs to verify sizes and contents
print(paste("Main term:", main_term))
print("Subterm matches (first 5 rows):")
print(head(subterm_matches_df, 5))
print("Exclusion matches (first 5 rows):")
print(head(exclusion_matches_df, 5))
# Update candidate identification
medical_combined <- medical_combined %>%
mutate(candidate = case_when(
grepl(main_term, term_aurum, ignore.case = TRUE) & num_subterms_found == 0 & num_exclusions_found == 0 ~ 1,
grepl(main_term, term_aurum, ignore.case = TRUE) & num_subterms_found > 0 & num_exclusions_found == 0 ~ 1,
TRUE ~ candidate
))
# Final check to remove candidates if exclusions are present
medical_combined <- medical_combined %>%
mutate(candidate = ifelse(candidate == 1 & num_exclusions_found > 0, 0, candidate))
}