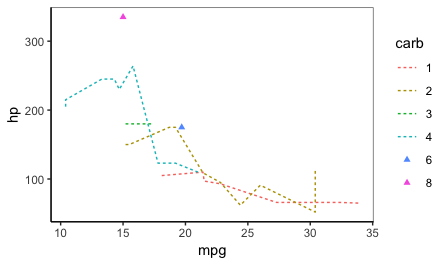
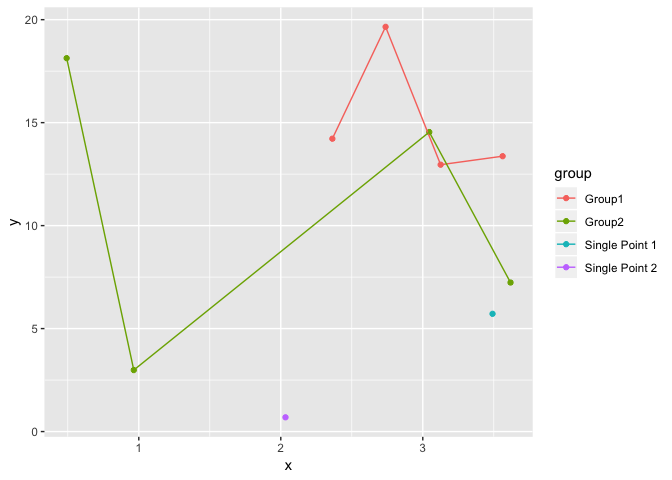
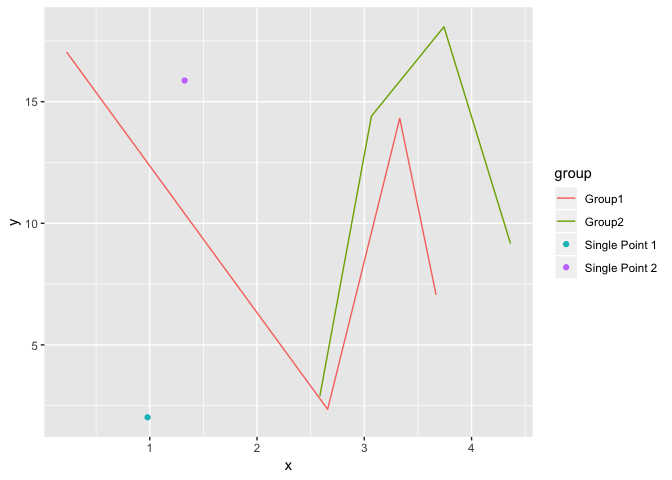
Below is a function using tidy evaluation that's intended to work on any data frame to plot singleton groups as points and non-singletons as lines only.
library(tidyverse)
plot_func = function(data, x, y, group.var, sort.singletons=TRUE,
shape=16, linetype="solid") {
# Quote arguments
x=enquo(x)
y=enquo(y)
group.var = enquo(group.var)
# Mark singleton groups
data = data %>%
group_by(!!group.var) %>%
mutate(single = n()==1) %>%
ungroup
# If sort.singletons (the default), place singleton groups after non-singletons in legend
if(sort.singletons) {
data = data %>%
arrange(single, !!group.var) %>%
mutate(!!group.var := factor(!!group.var, levels=unique(!!group.var)))
}
# Generate a named logical vector marking singleton groups
# Use this for generating the appropriate legend keys
single.groups = data %>%
group_by(!!group.var) %>%
slice(1) %>%
select(!!group.var, single) %>%
deframe()
# Generate legend key symbols based on single.groups vector we just created
shapes = single.groups %>% ifelse(., shape, NA_integer_)
lines = single.groups %>% ifelse(., "blank", linetype)
data %>%
ggplot(aes(x = !!x, y = !!y, color = !!group.var)) +
geom_line(linetype=linetype) +
geom_point(aes(shape=single)) +
scale_shape_manual(values=c(`FALSE`=NA, `TRUE`=shape)) +
guides(shape=FALSE,
colour=guide_legend(override.aes=list(shape=shapes,
linetype=lines)))
}
Now, run the function on various data frames:
plot_func(dat, x, y, group)
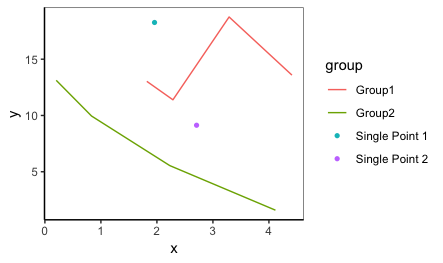
plot_func(iris, Petal.Width, Petal.Length, Species)
plot_func(iris %>% slice(c(1:50, 51, 101:150)),
Petal.Width, Petal.Length, Species)
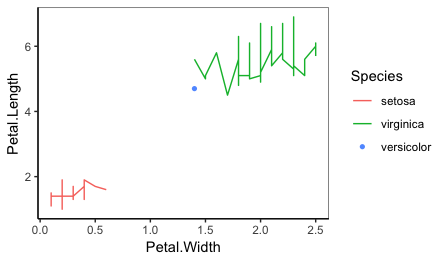
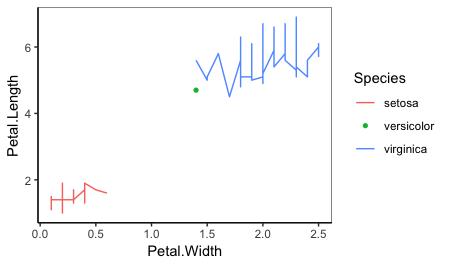
plot_func(iris %>% slice(c(1:50, 51, 101:150)),
Petal.Width, Petal.Length, Species, sort.singletons=FALSE)
plot_func(mtcars %>% mutate(carb=factor(carb)), mpg, hp, carb,
shape=17, linetype="22")