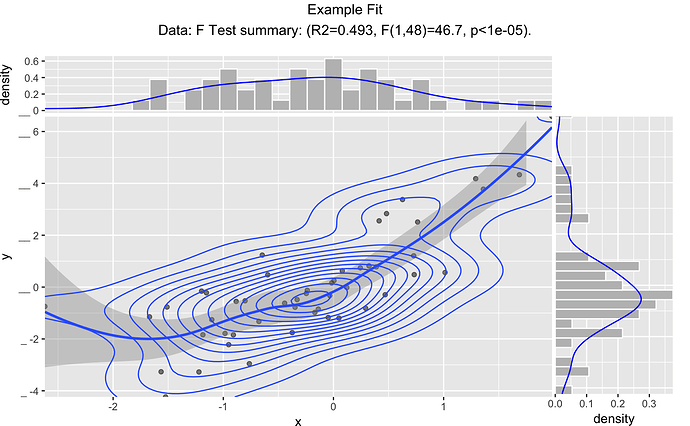
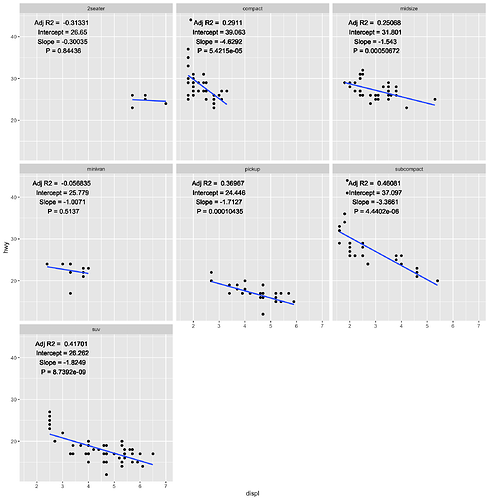
Thanks for your reply! It's very useful.
I tried to rewrite the code to create functions that would take data, xvar, yvar and group (for faceting) as arguments. However, there is something wrong with my syntax in both examples.
- single graph example
ggplotRegression <- function(dat, xvar, yvar){
require(ggplot2)
fit <- lm(yvar~xvar, dat)
ggplot(fit$model, aes_string(x = names(fit$model)[2], y = names(fit$model)[1])) +
geom_point() +
stat_smooth(method = "lm", col = "red") +
labs(title = paste("Adj R2 = ",signif(summary(fit)$adj.r.squared, 5),
"Intercept =",signif(fit$coef[[1]],5 ),
" Slope =",signif(fit$coef[[2]], 5),
" P =",signif(summary(fit)$coef[2,4], 5)))
}
ggplotRegression(Sepal.Length, Petal.Width, iris)
- facet graph example
library(tidyverse)
facetRegression <- function(dat, xvar, yvar, group) {
xvar <- enquo(xvar)
yvar <- enquo(yvar)
group <- enquo(group)
dat %>%
nest(-!!group) %>%
mutate(model = map(data, ~ lm(!!xvar~!!yvar, data = .x)),
adj.r.squared = map_dbl(model, ~ signif(summary(.x)$adj.r.squared, 5)),
intercept = map_dbl(model, ~ signif(.x$coef[[1]],5)),
slope = map_dbl(model, ~ signif(.x$coef[[2]], 5)),
pvalue = map_dbl(model, ~ signif(summary(.x)$coef[2,4], 5))
) %>%
select(-data, -model) %>%
left_join(dat) %>%
ggplot(aes_(substitute(xvar), substitute(yvar))) +
geom_point() +
geom_smooth(se = FALSE, method = "lm") +
facet_wrap(~!!group) +
geom_text(aes(3, 40, label = paste("Adj R2 = ", adj.r.squared, "\n",
"Intercept =",intercept, "\n",
"Slope =", slope, "\n",
"P =", pvalue)))
}
facetRegression(mpg, displ, hwy, drv)
These are probably very naive attempts, but I'm not very experienced in writing functions. In the past, I wrote some non-elegant functions in base R but after switching to tidyverse I got confused with tidy evaluation, so now I'm not even sure where I need to use it and where I'm fine with the standard way of writing function (I think I don't need the tidy eval in the single graph example, right?).
There is clearly something wrong in the way I specify the model equation within both functions. I tried to write the equation in a couple of different ways, e.g.,
lm(yvar~xvar, dat)
with(dat, lm(yvar~xvar))
lm(dat[[yvar]]~dat[[xvar]])
...but no success 
For the facet example, I also tried to separate the code into two steps i) data preparation and ii) ggplot plotting, but was not able to get the function working properly for either piece of the code.
I thought about the strategy for positioning the text in each facet. I think it should be possible to get ymax and xmin for each drv group in the data preparation phase and then use it with some reasonable offset within geom_text(), but typing the correct syntax it is clearly beyond my current programming skills 
I would appreciate help with the code above, but I would be extremely grateful for some "philosophical" discussion on how to approach building functions like these step by step. I don't have CS background (I come from the field of biology) and use R to analyze my biological data. I often find myself in the situation when I get some analyses and graphs done, but as the project grows I start to get lost in my code. I try to simplify it (e.g., by writing a function) but then get stuck at some programming obstacle. I then spend a lot of time fiddling with the code and often don't get over the obstacle because I cannot come up with the right syntax. I guess my problem is that I want to achieve too complex things for my level of programming skills, but how do I know what's the right level of complexity is. I want to be falling in the pit of programming success