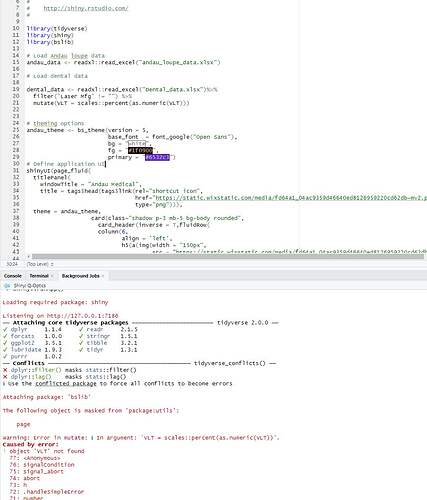
You are right! I asked my boss if he recalled having a column named VLT in the database and he said yup. The previous programmer didn't give him the access to the code files, so I think I'm stuck with the old files and I'm doing my best to get it fixed, hopefully the best version! I appreciate all the help while trying to learn at the same time. 
Alright, here's the output...
andau_data
=> shiny::runApp()
Loading required package: shiny
Listening on http://127.0.0.1:4860
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package to force all conflicts to become errors
Attaching package: 'bslib'
The following object is masked from 'package:utils':
page
structure(list(`Andau Frame` = c("Bolle", "ProGear", "MOS", "MOS-W",
"Blues LG", "Blues MD", "Indie LG", "Indie MD", "Soul LG", "Soul MD",
"Jazz"), `Innovative Optics Insert` = c("IVR", "IVL", "IVL.R",
"IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R",
"IVR")), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-11L))
structure(list(`Laser Mfg` = c("Biolase", "Biolase", "DeKa",
"Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Millennium",
"Denmat", "AMD", "AMD", "AMD", "AMD", "Brasseler", "CAO", "Clincians Choice",
"Denmat", "THOR", "THOR", "THOR", "Zolar", "Biolase", "Biolase",
"Biolase", "King Medical", "Sirona Dentsply", "Sirona Dentsply",
"Ultradent", "Zolar", "Ivoclar Vivadent", "Convergent Dental",
"DeKa", "DeKa", "GPT Dental", "Lightscalpel", "Lightscalpel",
"Zap Lasers"), `Laser Model` = c("Waterlase iPlus", "Waterlase Express",
"SmartPerio ", "LightWalker AT", "LightWalker AT S", "Lightwalker ST-E",
"SkyPulse Endo", "SkyPulse Versa", "NightLase", "PerioLase MVP-7",
"NVPro3", "Picasso+", "Picasso+ Lite", "Clarion", "Picasso Clario",
"BLU Dental Microlaser", "Precise LTM", "Clincians Choice", "Sol",
"810nm Infra Red Single Laser Dental Probe", "810nm 1W laser cluster (5 x 200mW)",
"810nm 200mW single laser probe", "Photon dental diode laser",
"Epic X", "Epic Pro", "Epic Hygiene", "Beamer STL 980", "SiroLaser Advance Plus",
"Siro Blue", "Gemini Laser", "Photon Plus Dental Diode Laser",
"Odyssey Diode", "Solea Laser", "Smart US-20D CO2 Laser", "SmartXide Ultraspeed2 CO2 Laser",
"DENTA CO2", "LS-1005", "LS-2010", "Softlase"), Wavelengths = c("2780nm",
"2780nm", "1064nm", "1064nm / 2940nm", "1064nm / 2940nm", "2940nm",
"2940nm", "2940nm", "2940nm", "1064nm", "808nm (OD 4)", "800-820nm OD 5",
"800-820nm OD 5", "800-820nm OD 5", "810nm", "810nm", "810nm",
"808nm", "810nm", "810nm OD 3", "810nm OD 4", "810nm OD 3", "810nm",
"940nm +/- 10nm", "940nm +/- 20nm, 980nm +/- 20nm", "980nm +/- 10nm",
"980nm", "660nm +/- 5nm, 970nm -10/+15nm", "440-450nm OD5, 655-665nm OD1, 960-985nm OD5",
"810nm / 980nm", "980nm", "810nm +/- 20nm", "9,300nm", "10,600nm",
"10,600nm", "10600nm", "10600nm", "10600nm", "630-670nm (OD 0.7), 800-980nm (OD 3.6)"
), Lens = c("Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1",
"Gi1", "Gi1", "Gi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1",
"Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi17", "Pi17",
"Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi19",
"Pi19", "Pi19", "Pi19", "Pi19", "Pi19", "Pi17")), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -39L))
structure(list(`Andau Frame` = c("Bolle", "ProGear", "MOS", "MOS-W",
"Blues LG", "Blues MD", "Indie LG", "Indie MD", "Soul LG", "Soul MD",
"Jazz"), `Innovative Optics Insert` = c("IVR", "IVL", "IVL.R",
"IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R",
"IVR")), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-11L))
structure(list(`Laser Mfg` = c("Biolase", "Biolase", "DeKa",
"Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Millennium",
"Denmat", "AMD", "AMD", "AMD", "AMD", "Brasseler", "CAO", "Clincians Choice",
"Denmat", "THOR", "THOR", "THOR", "Zolar", "Biolase", "Biolase",
"Biolase", "King Medical", "Sirona Dentsply", "Sirona Dentsply",
"Ultradent", "Zolar", "Ivoclar Vivadent", "Convergent Dental",
"DeKa", "DeKa", "GPT Dental", "Lightscalpel", "Lightscalpel",
"Zap Lasers"), `Laser Model` = c("Waterlase iPlus", "Waterlase Express",
"SmartPerio ", "LightWalker AT", "LightWalker AT S", "Lightwalker ST-E",
"SkyPulse Endo", "SkyPulse Versa", "NightLase", "PerioLase MVP-7",
"NVPro3", "Picasso+", "Picasso+ Lite", "Clarion", "Picasso Clario",
"BLU Dental Microlaser", "Precise LTM", "Clincians Choice", "Sol",
"810nm Infra Red Single Laser Dental Probe", "810nm 1W laser cluster (5 x 200mW)",
"810nm 200mW single laser probe", "Photon dental diode laser",
"Epic X", "Epic Pro", "Epic Hygiene", "Beamer STL 980", "SiroLaser Advance Plus",
"Siro Blue", "Gemini Laser", "Photon Plus Dental Diode Laser",
"Odyssey Diode", "Solea Laser", "Smart US-20D CO2 Laser", "SmartXide Ultraspeed2 CO2 Laser",
"DENTA CO2", "LS-1005", "LS-2010", "Softlase"), Wavelengths = c("2780nm",
"2780nm", "1064nm", "1064nm / 2940nm", "1064nm / 2940nm", "2940nm",
"2940nm", "2940nm", "2940nm", "1064nm", "808nm (OD 4)", "800-820nm OD 5",
"800-820nm OD 5", "800-820nm OD 5", "810nm", "810nm", "810nm",
"808nm", "810nm", "810nm OD 3", "810nm OD 4", "810nm OD 3", "810nm",
"940nm +/- 10nm", "940nm +/- 20nm, 980nm +/- 20nm", "980nm +/- 10nm",
"980nm", "660nm +/- 5nm, 970nm -10/+15nm", "440-450nm OD5, 655-665nm OD1, 960-985nm OD5",
"810nm / 980nm", "980nm", "810nm +/- 20nm", "9,300nm", "10,600nm",
"10,600nm", "10600nm", "10600nm", "10600nm", "630-670nm (OD 0.7), 800-980nm (OD 3.6)"
), Lens = c("Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1",
"Gi1", "Gi1", "Gi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1",
"Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi17", "Pi17",
"Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi19",
"Pi19", "Pi19", "Pi19", "Pi19", "Pi19", "Pi17")), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -39L))
dental_data
=> shiny::runApp()
Loading required package: shiny
Listening on http://127.0.0.1:7544
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package to force all conflicts to become errors
structure(list(`Andau Frame` = c("Bolle", "ProGear", "MOS", "MOS-W",
"Blues LG", "Blues MD", "Indie LG", "Indie MD", "Soul LG", "Soul MD",
"Jazz"), `Innovative Optics Insert` = c("IVR", "IVL", "IVL.R",
"IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R", "IVL.R",
"IVR")), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-11L))
structure(list(`Laser Mfg` = c("Biolase", "Biolase", "DeKa",
"Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Millennium",
"Denmat", "AMD", "AMD", "AMD", "AMD", "Brasseler", "CAO", "Clincians Choice",
"Denmat", "THOR", "THOR", "THOR", "Zolar", "Biolase", "Biolase",
"Biolase", "King Medical", "Sirona Dentsply", "Sirona Dentsply",
"Ultradent", "Zolar", "Ivoclar Vivadent", "Convergent Dental",
"DeKa", "DeKa", "GPT Dental", "Lightscalpel", "Lightscalpel",
"Zap Lasers"), `Laser Model` = c("Waterlase iPlus", "Waterlase Express",
"SmartPerio ", "LightWalker AT", "LightWalker AT S", "Lightwalker ST-E",
"SkyPulse Endo", "SkyPulse Versa", "NightLase", "PerioLase MVP-7",
"NVPro3", "Picasso+", "Picasso+ Lite", "Clarion", "Picasso Clario",
"BLU Dental Microlaser", "Precise LTM", "Clincians Choice", "Sol",
"810nm Infra Red Single Laser Dental Probe", "810nm 1W laser cluster (5 x 200mW)",
"810nm 200mW single laser probe", "Photon dental diode laser",
"Epic X", "Epic Pro", "Epic Hygiene", "Beamer STL 980", "SiroLaser Advance Plus",
"Siro Blue", "Gemini Laser", "Photon Plus Dental Diode Laser",
"Odyssey Diode", "Solea Laser", "Smart US-20D CO2 Laser", "SmartXide Ultraspeed2 CO2 Laser",
"DENTA CO2", "LS-1005", "LS-2010", "Softlase"), Wavelengths = c("2780nm",
"2780nm", "1064nm", "1064nm / 2940nm", "1064nm / 2940nm", "2940nm",
"2940nm", "2940nm", "2940nm", "1064nm", "808nm (OD 4)", "800-820nm OD 5",
"800-820nm OD 5", "800-820nm OD 5", "810nm", "810nm", "810nm",
"808nm", "810nm", "810nm OD 3", "810nm OD 4", "810nm OD 3", "810nm",
"940nm +/- 10nm", "940nm +/- 20nm, 980nm +/- 20nm", "980nm +/- 10nm",
"980nm", "660nm +/- 5nm, 970nm -10/+15nm", "440-450nm OD5, 655-665nm OD1, 960-985nm OD5",
"810nm / 980nm", "980nm", "810nm +/- 20nm", "9,300nm", "10,600nm",
"10,600nm", "10600nm", "10600nm", "10600nm", "630-670nm (OD 0.7), 800-980nm (OD 3.6)"
), Lens = c("Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1",
"Gi1", "Gi1", "Gi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1",
"Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi17", "Pi17",
"Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi19",
"Pi19", "Pi19", "Pi19", "Pi19", "Pi19", "Pi17")), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -39L))
Attaching package: 'bslib'
The following object is masked from 'package:utils':
page
structure(list(`Laser Mfg` = c("Biolase", "Biolase", "DeKa",
"Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Fotona", "Millennium",
"Denmat", "AMD", "AMD", "AMD", "AMD", "Brasseler", "CAO", "Clincians Choice",
"Denmat", "THOR", "THOR", "THOR", "Zolar", "Biolase", "Biolase",
"Biolase", "King Medical", "Sirona Dentsply", "Sirona Dentsply",
"Ultradent", "Zolar", "Ivoclar Vivadent", "Convergent Dental",
"DeKa", "DeKa", "GPT Dental", "Lightscalpel", "Lightscalpel",
"Zap Lasers"), `Laser Model` = c("Waterlase iPlus", "Waterlase Express",
"SmartPerio ", "LightWalker AT", "LightWalker AT S", "Lightwalker ST-E",
"SkyPulse Endo", "SkyPulse Versa", "NightLase", "PerioLase MVP-7",
"NVPro3", "Picasso+", "Picasso+ Lite", "Clarion", "Picasso Clario",
"BLU Dental Microlaser", "Precise LTM", "Clincians Choice", "Sol",
"810nm Infra Red Single Laser Dental Probe", "810nm 1W laser cluster (5 x 200mW)",
"810nm 200mW single laser probe", "Photon dental diode laser",
"Epic X", "Epic Pro", "Epic Hygiene", "Beamer STL 980", "SiroLaser Advance Plus",
"Siro Blue", "Gemini Laser", "Photon Plus Dental Diode Laser",
"Odyssey Diode", "Solea Laser", "Smart US-20D CO2 Laser", "SmartXide Ultraspeed2 CO2 Laser",
"DENTA CO2", "LS-1005", "LS-2010", "Softlase"), Wavelengths = c("2780nm",
"2780nm", "1064nm", "1064nm / 2940nm", "1064nm / 2940nm", "2940nm",
"2940nm", "2940nm", "2940nm", "1064nm", "808nm (OD 4)", "800-820nm OD 5",
"800-820nm OD 5", "800-820nm OD 5", "810nm", "810nm", "810nm",
"808nm", "810nm", "810nm OD 3", "810nm OD 4", "810nm OD 3", "810nm",
"940nm +/- 10nm", "940nm +/- 20nm, 980nm +/- 20nm", "980nm +/- 10nm",
"980nm", "660nm +/- 5nm, 970nm -10/+15nm", "440-450nm OD5, 655-665nm OD1, 960-985nm OD5",
"810nm / 980nm", "980nm", "810nm +/- 20nm", "9,300nm", "10,600nm",
"10,600nm", "10600nm", "10600nm", "10600nm", "630-670nm (OD 0.7), 800-980nm (OD 3.6)"
), Lens = c("Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1", "Gi1",
"Gi1", "Gi1", "Gi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1",
"Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi1", "Pi17", "Pi17",
"Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi17", "Pi19",
"Pi19", "Pi19", "Pi19", "Pi19", "Pi19", "Pi17")), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -39L))