Hi,
I have a large dataset of adjacencies. By this I mean data like:-
links=data.frame(
source=c("K", "A","A", "A", "A", "A","J", "B", "B", "C", "C", "D","I" ,"K", "A", "A"),
target= c("P", "B","B", "C", "D", "J", "A","E", "F", "G", "H", "I","I" , "L", "G", "C")
In reality my data has tens of thousands of adjacencies and when networked it will create many distinct/separate networks of various sizes. The example data above creates two distinct networks - my data will have hundreds of distinct non-linking networks.
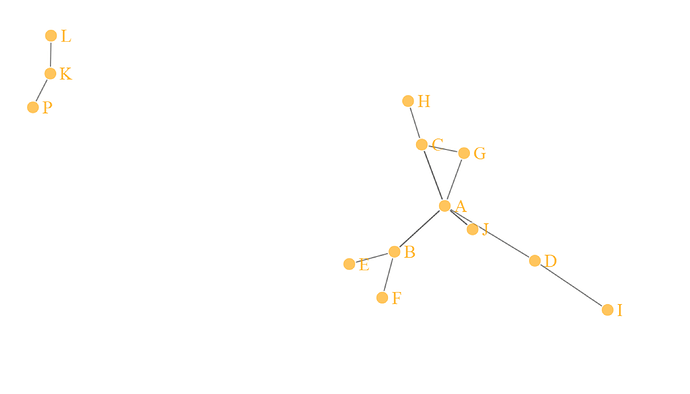
Just picking a basic network function ('simpleNetwork') on the above data gives the following visualisation.
So on this simple data, I'm after an output dataset that has a network ID - like the mocked-up table below. I'm less interested in the visualisation.
Network ID source target
1 L K
1 K P
2 H C
2 C G
2 C A
etc
This table shows me which adjacencies are in which network and will allow me to build network level metrics such as network size etc and overlay some other data I have - and then I can select certain ones of interest to visualise.
Is there an R function/package that will get me this kind of table?
regards,
Andy