Hi,
I am trying to plot eleven lines showing contaminant concentration data for eleven sample sites over time (i.e, one line per site).
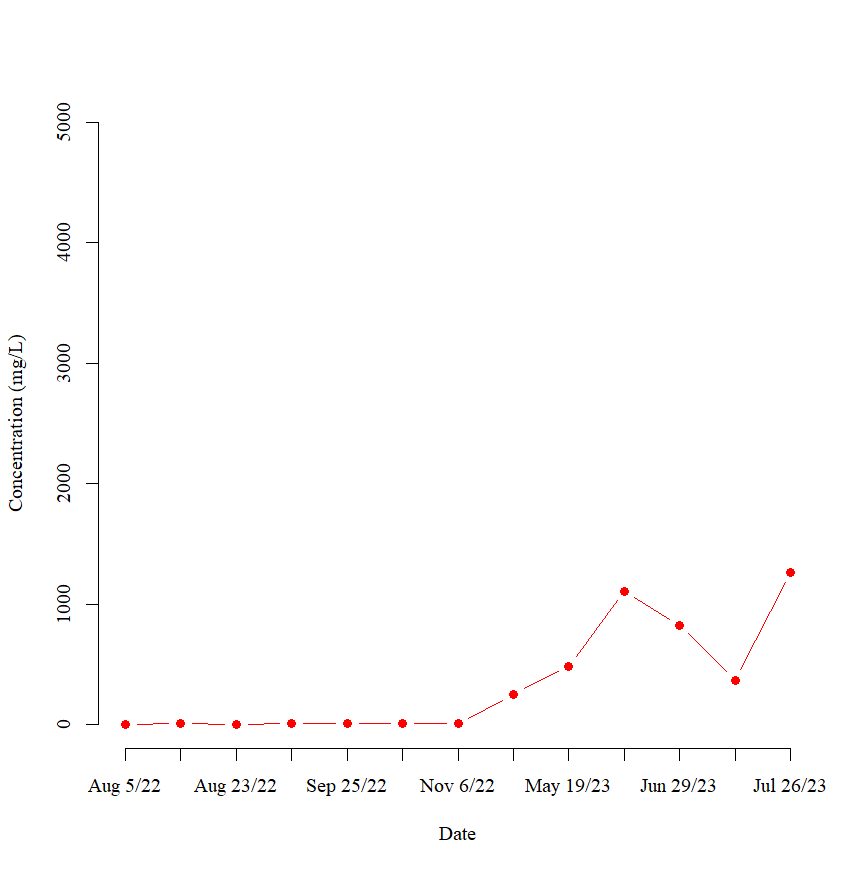
To do this, I started by plotting the concentrations from the first site using the code:
plot(TSSConc.dat$C1In1, type = "b", xlab = "Date", ylab = "Concentration (mg/L)", frame = FALSE, pch = 19, col = "red", xaxt = "n", family = "A", ylim = c(0,5000))
This code returns the following plot:
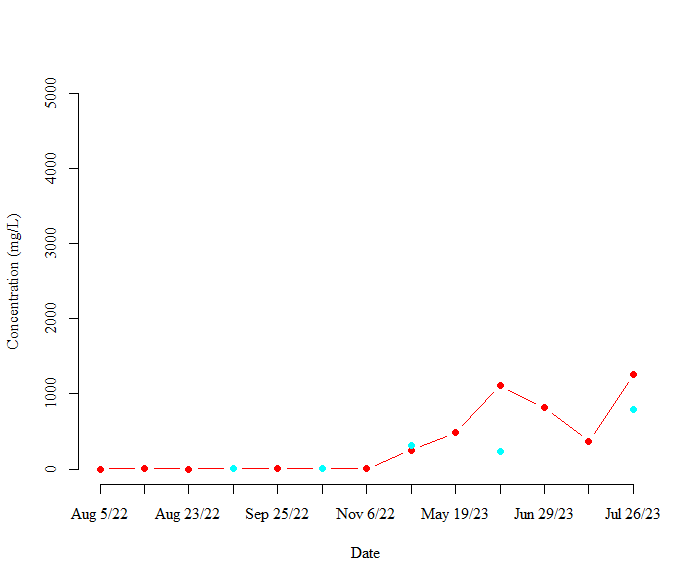
I then used the lines function to add my next sample site to using the code
lines(TSSConc.dat$C2In, type = "b", col = "cyan", lty = 1, lwd = 2, pch = 16)
This code returns the following plot:
As shown in the above plot, the cyan points are not connected by any lines. I assume that this is because there are missing values for this sample site.
As I continue to add sites, the lines do not draw correctly for sites with missing values. Is there a way to get R to bypass the missing value and draw a line to the next value?
TIA