Good evening,
I have been trying to add a subscript for (H20) to the legend using scale_color_manual. I had to create another aes mapping to see and manually alter the legend. However, I am unable so far to add a subscript. I have been using this code:
geom_point(aes(colour = "Blank (50 µM of dH20)"))+
scale_color_manual(name = "Final concentrations and volumes",
values = c(expression(paste( "Blank" ~ "(", 50 *µL* "of", dH[2],0, ")" = "#B2182B"))),
expression(paste( "Vehicle - Vehicle"~ "(", 10 *µL* "of", dH[2],0 , "+", 10 *µL* "of", dH[2],0, ")" = "#D6604D")),
expression(paste( "Vehicle -Nicotinic Acid" ~ "(", 10 *µL* "of", dH[2],0 , "+", 10 *µL* "of", 1 *mM* ")" = "#F4A582")),
expression(paste( "Sp-Vehicle" ~ "(", 10 *µL* "of", 5 *µM* "+", 10 *µL* "of", dH[2], 0, ")" = "#FDDBC7")),
expression(paste( "Sp-Nicotinic Acid" ~ "(", 10 *µL* "of", 5 *µM* "+", 10 *µL* "of", 1 *mM* ")" = "#D1E5F0")))+
But, I am encountering the following error:
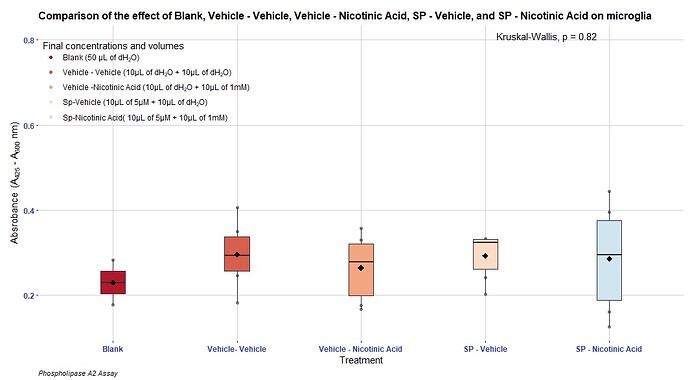
I used "=" to order ggplot2 to produce the exact same colours as they are presented within the graph:
As I did previously:
scale_color_manual(name = "Final concentrations and volumes",
labels = c( "Blank (50 µL of dH20)", "Vehicle - Vehicle (10µL of dH20 + 10µL of dH20)", "Vehicle -Nicotinic Acid (10µL of dH20 + 10µL of 1mM)", "Sp-Vehicle (10µL of 5µM + 10µL of dH20)", "Sp-Nicotinic Acid( 10µL of 5µM + 10µL of 1mM)"),
values = c("Blank (50 µL of dH20)" = "#B2182B", "Vehicle - Vehicle (10µL of dH20 + 10µL of dH20)" = "#D6604D", "Vehicle -Nicotinic Acid (10µL of dH20 + 10µL of 1mM)" = "#F4A582", "Sp-Vehicle (10µL of 5µM + 10µL of dH20)"= "#FDDBC7", "Sp-Nicotinic Acid(10µL of 5µM + 10µL of 1mM)"= "#D1E5F0"))+
I do not know where I am doing mistakes.
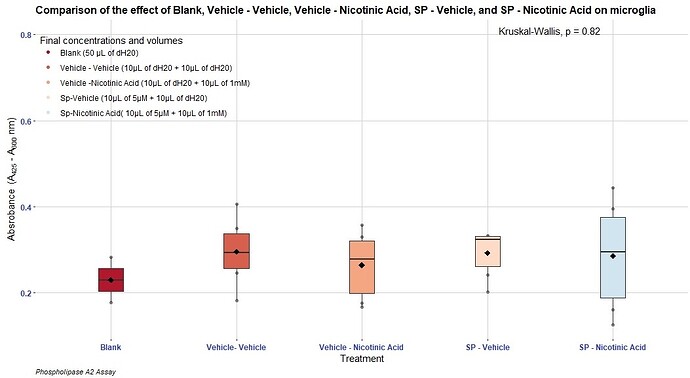
That is the entire code used to produce the above graph:
PhospholipaseA2NOPo1<-structure(list(Drug = structure(c(1L, 1L, 5L, 5L, 5L, 5L, 5L,
5L, 4L, 4L, 4L, 4L, 4L, 4L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L,
2L, 2L, 2L), .Label = c("Blank", "SP.N", "SP.V", "V.N", "V.V"
), class = "factor"), Absorbance = c(0.178155098, 0.281888047,
0.315468607, 0.395427286, 0.443773761, 0.274327232, 0.126408569,
0.160375886, 0.332019768, 0.328779358, 0.31918001, 0.202597307,
0.241052828, 0.332138474, 0.356656925, 0.329787758, 0.291912398,
0.264823437, 0.167194165, 0.176451481, 0.406376062, 0.28900749,
0.298281464, 0.181890255, 0.245723697, 0.350279692)), class = "data.frame", row.names = c(NA,
-26L))
library(tidyverse)
library(ggplot2)
library(ggpubr)
library(RColorBrewer)
library(tidyr)
library(readr)
library(cowplot)
ggplot(PhospholipaseA2NOPo1, aes( x = Drug, y = Absorbance))+
labs(x = expression(paste("Treatment")))+
ylab(expression(paste("Absrobance " ~ "(", A[425], " - ", A[600] ~ nm, ")"))) +
labs(title = "Comparison of the effect of Blank, Vehicle - Vehicle, Vehicle - Nicotinic Acid, SP - Vehicle, and SP - Nicotinic Acid on microglia ",
caption = "Phospholipase A2 Assay")+
geom_point(aes(colour = "Blank (50 µM of dH20)"))+
scale_color_manual(name = "Final concentrations and volumes",
labels = c( "Blank (50 µL of dH20)", "Vehicle - Vehicle (10µL of dH20 + 10µL of dH20)", "Vehicle -Nicotinic Acid (10µL of dH20 + 10µL of 1mM)", "Sp-Vehicle (10µL of 5µM + 10µL of dH20)", "Sp-Nicotinic Acid( 10µL of 5µM + 10µL of 1mM)"),
values = c("Blank (50 µL of dH20)" = "#B2182B", "Vehicle - Vehicle (10µL of dH20 + 10µL of dH20)" = "#D6604D", "Vehicle -Nicotinic Acid (10µL of dH20 + 10µL of 1mM)" = "#F4A582", "Sp-Vehicle (10µL of 5µM + 10µL of dH20)"= "#FDDBC7", "Sp-Nicotinic Acid(10µL of 5µM + 10µL of 1mM)"= "#D1E5F0"))+
stat_compare_means(label.y = 0.80,
label.x = 4.25)+
scale_x_discrete(labels = c("Blank", "Vehicle- Vehicle", "Vehicle - Nicotinic Acid", "SP - Vehicle", "SP - Nicotinic Acid"))+
geom_boxplot(
fill=c("#B2182B", "#D6604D", "#F4A582", "#FDDBC7", "#D1E5F0"),
width=0.2,notch = FALSE)+
stat_summary(fun=mean, colour="black", geom="point",
shape=18, size=3) +
theme_bw() +
theme(panel.grid.major = element_line(colour = "#d3d3d3"),
panel.grid.minor = element_blank(),
panel.border = element_blank(), panel.background = element_blank(),
plot.title = element_text(size = 14, face = "bold"))+
theme(axis.text.x = element_text(face="bold", color="#3142b0",
size=8.9, angle=0),
axis.text.y = element_text(face="bold", color="#3142b0",
size=8.9, angle=0))+
theme(
plot.title = element_text(size = 12, face = "bold"),
plot.caption = element_text(hjust = 0, size = 8, face = "italic")
)+
scale_linetype_manual('Legend',values='solid')+
scale_shape_manual('',values = 18)+
theme(legend.spacing.y = unit(0.01, "cm"))+
theme(legend.position = c(0, .97),
legend.justification = c("left", "top"),
legend.background = element_rect(colour = NA, fill = NA))
And this is the : DataUsed