Hello everyone,
Hello to all of you,
I'm making an interactive R map with the Leaflet package, the application domain is related to zoology with the distribution of the Hedgehogs of the World.
Luckily I have already found some well done tutorials on the internet to train myself and produce the result.
Nevertheless, to make it perfect I still have a problem with one thing of great importance.
Here's my code
# - - - Project Map Hedgehogs
library(shiny)
library(shinythemes)
library(leaflet)
library(leaflet.providers)
library(dplyr)
library(maps)
library(RColorBrewer)
library(leaflet.extras)
library(rworldxtra)
library(raster)
library(sf)
library(tidyverse)
# 0] Load data
Data_Herisson <- read.csv("XXXXX/Documents/Question hérisson.csv")
# 1] Color from a factor which is " Genus Name "
Genus_names <- Data_Herisson$genus_name %>% unique()
# Palette de couleurs
library(RColorBrewer)
brewer.pal(n = 3, name = "Paired")
Color <- c("blue","red","green")
pal <- colorFactor(Color,domain = Genus_names)
# [FINAL] Afficher la carte
Map_Herisson<-leaflet() %>%
addProviderTiles("Esri.WorldImagery") %>%
addCircleMarkers(data = Data_Herisson,
lat = ~latitude,
lng = ~longitude,
color = ~pal(Data_Herisson$genus_name),
stroke = FALSE,
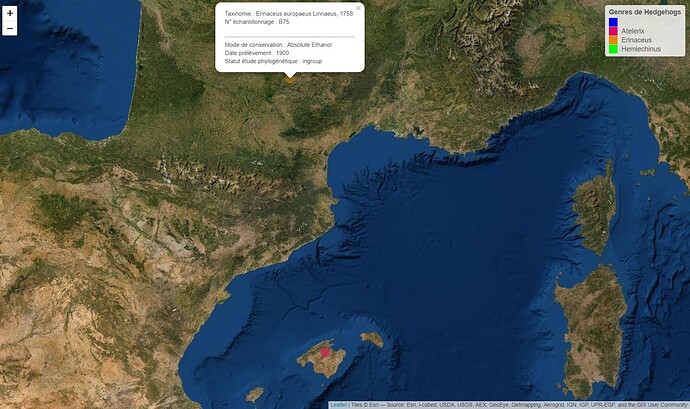
popup = paste("Taxinomie : ", Data_Herisson$complete_name,
"<br>",
"N° échantillonnage : ", Data_Herisson$specimen_code,
"<br>","<br>","<hr>",
"Mode de conservation : ", Data_Herisson$preservation_method,
"<br>",
"Date prélèvement : ", Data_Herisson$date_collected,
"<br>",
"Statut étude phylogénétique : ", Data_Herisson$phylogeny_status),
label = ~as.character(name),
clusterOptions = markerClusterOptions(),
fillOpacity = 0.5)
# Ajout Légende des Genres à la Carte
Map_Herisson %>% addLegend(data = Data_Herisson,
position = "topright",
pal = pal,
values = Data_Herisson$genus_name,
title = "Genres de Brentidae",
opacity = 1)
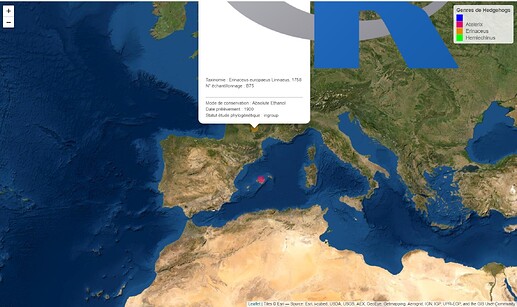
Voici ce que j'obtiens ce qui est déjà très satisfaisant :
My question is the following, how can I display in the popup a photo of each specimen?
It would be possible to use a link to a photo of the taxon, but that would break the aesthetic and interactive interest of the thing.
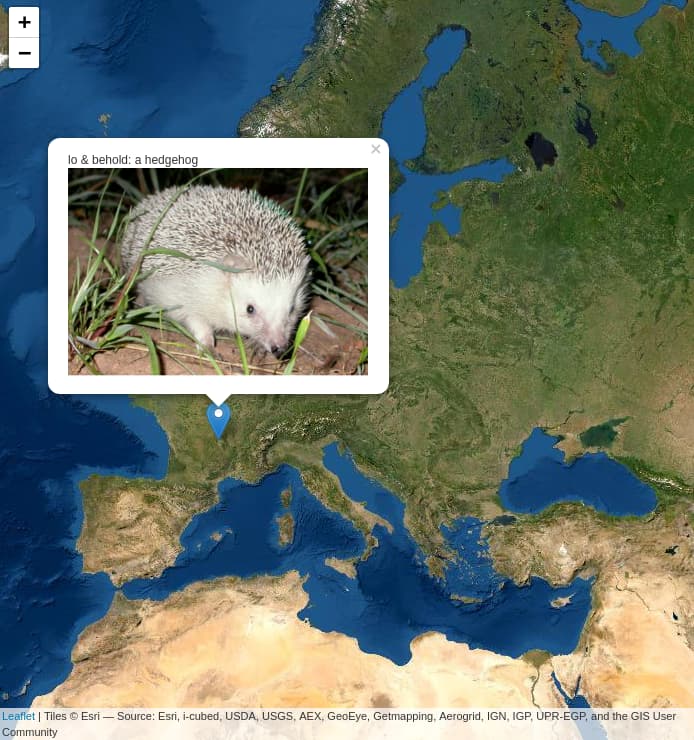
I have already seen and understood how to add a photo that is displayed for each marker (Source : Add a pop-up with an image to a leaflet map in R) but I can't figure out how to do it knowing that my real dataset is much larger .... ![]()
If this works, the result would be truly wonderful ! ![]()
Thak you in advance, wishing you a great day