Hello,
Say I have a matrix of size 100x2
X = ( (x_1, y_1),
(x_2, y_2),
...,
(x_100, y_100))
Each entry (x_i, y_i) is a point of the unit circle.
How can I get a 2D plot of this matrix?
Hello,
Say I have a matrix of size 100x2
X = ( (x_1, y_1),
(x_2, y_2),
...,
(x_100, y_100))
Each entry (x_i, y_i) is a point of the unit circle.
How can I get a 2D plot of this matrix?
I'm not sure if I understand you correctly because you are not providing a REPRoducible EXample (reprex). Is this what you are trying to do?
library(ggplot2)
# Made up X matrix
X <- matrix(1:100, nrow = 50, ncol = 2)
X <- as.data.frame(X)
names(X) <- c("x", "y")
head(X)
#> x y
#> 1 1 51
#> 2 2 52
#> 3 3 53
#> 4 4 54
#> 5 5 55
#> 6 6 56
# Plot
ggplot(X, aes(x = x, y = y)) +
geom_point()
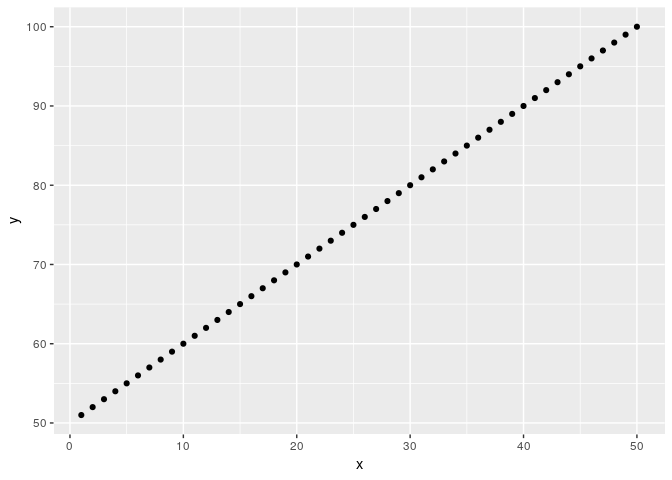
Created on 2019-04-07 by the reprex package (v0.2.1.9000)
I have 100 couples (x_i, y_i) in a matrix such that x_i^2 + y_i^2 = 1 and I want to plot them.
Well, my code above is plotting (x, y) points, you can modify the code to do the same with your own matrix, if you want more specific help, please provide a reproducible example.
If you've never heard of a reprex before, you might want to start by reading this FAQ:
If you have the coordinates of the points you want to plot in two columns of a matrix, you can simply use the plot function on that matrix. You don't need to use ggplot here. But of course, you can use it.
See below:
r <- 1
theta <- seq(from = -pi,
to = pi,
length.out = 1e+3)
u <- (r * cos(x = theta))
v <- (r * sin(x = theta))
data_in_matrix <- cbind(u, v)
# base plot function
plot(data_in_matrix,
asp = 1)
# using ggplot2
library(ggplot2)
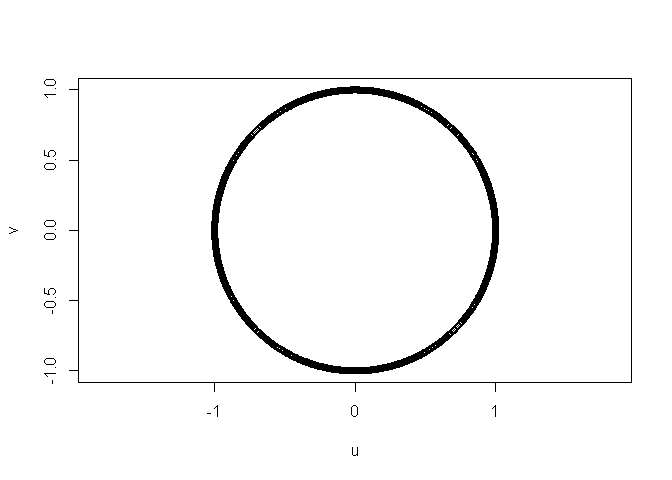
# quickplot
quickplot(x = u,
y = v,
data = as.data.frame(x = data_in_matrix))
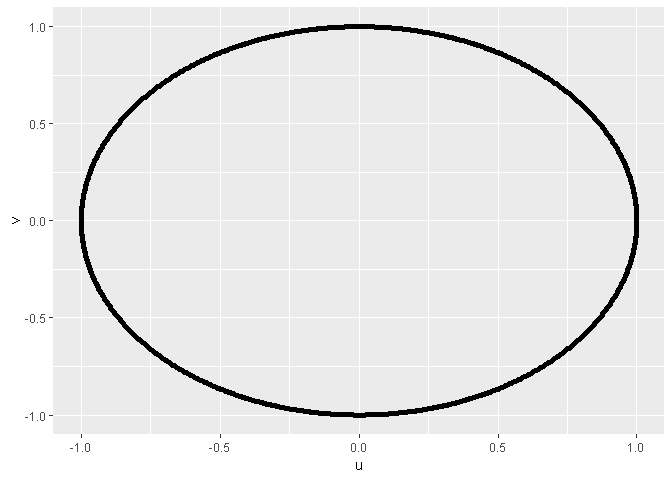
# ggplot
ggplot(data = as.data.frame(x = data_in_matrix),
mapping = aes(x = u,
y = v)) +
geom_point()

Created on 2019-04-07 by the reprex package (v0.2.1)
Thanks a lot, that was I was looking for.
Does it still work in dimension 3 e.g. for X = ( (x_1, y_1, z_1),
(x_2, y_2, z_2),
...,
(x_100, y_100, z_100))?
ggplot2 doesn't support 3D plotting but there are other options for this, like plotly for example.
library(plotly)
plot_ly(data = X, x = ~x, y = ~y, z = ~z, type = "scatter3d", mode = "markers")
Thanks.
How did you resize your plots?
I get an "ellipse":
instead of a circle because the width is 4 times the height.
I tried the code:
Y = simuSphere(3, 10000)
Y = as.data.frame(Y)
names(Y) <- c("x", "y", "z")
plot_ly(data = Y, x = x, y = y, z = z, type = "scatter3d", mode = "markers")
where simuSphere(3, 10000) gives me a matrix like 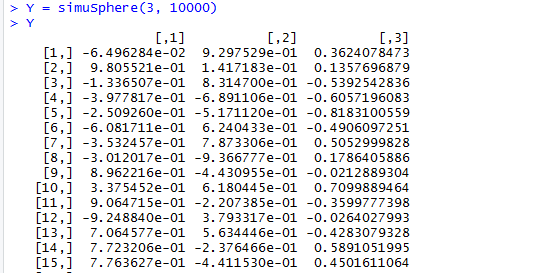
but I got a 2D figure:
Do you know why?
You are missing this symbol ~
plot_ly(data = Y, x = ~x, y = ~y, z = ~z, type = "scatter3d", mode = "markers")
To use same scale in both axes, use coord_fixed.
You may wish to take a look at this SO thread:
It's still a 2D plot even with the tilde
It's hard to know why without having a reproducible example, could you make one? I don't have access to the simuSphere() function, so I can't test your code.
Sure, here is the code:
norme2 = function(x){
return(sqrt(sum(x^2)))
}
simuNormale = function(n) {
u = runif(n)
v = runif(n)
x = sqrt(-2*log(u))*sin(2*pi*v)
return(x)
}
simuSphere = function(d, n){
X = matrix(nrow = n, ncol = d)
for(i in 1:n){
N = simuNormale(d)
X[i,] = N/norme2(N)
}
return(X)
}
I can't reproduce your issue, I get a 3D plot
library(plotly)
norme2 = function(x){
return(sqrt(sum(x^2)))
}
simuNormale = function(n) {
u = runif(n)
v = runif(n)
x = sqrt(-2*log(u))*sin(2*pi*v)
return(x)
}
simuSphere = function(d, n){
X = matrix(nrow = n, ncol = d)
for(i in 1:n){
N = simuNormale(d)
X[i,] = N/norme2(N)
}
return(X)
}
Y = simuSphere(3, 10000)
Y = as.data.frame(Y)
names(Y) <- c("x", "y", "z")
plot_ly(data = Y, x = ~x, y = ~y, z = ~z, type = "scatter3d", mode = "markers")
Weird, could it be a RStudio issue?
I've noticed that I don't get the plot for my graphics, I have to get them from Plots > save as image
I don't think so, try to run the code again on a fresh R session, Ctrl+Shift+F10
I did that and also tried updating the plot ly library and restarting my computer but it didn't change anything
Well, this seems like a different issue, I think you should ask this in a new topic (under #rstudio-ide category) and include all the relevant information to reproduce your problem including your R, RStudio and OS versions.
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.