Hello, first of all many thanks for your reply!
Here's a quick summary of my topic: This dissertation explores the non-food uses of honey from melipona bees in the Pinselli-Soyah-Sabouyah park in Guinea Conakry, focusing on the transmission of traditional knowledge. The study identified local Melipona species, documented traditional uses of honey, and assessed their cultural significance. Through 73 interviews in 16 localities, it was revealed that honey is used primarily as medicine, but also for ritual and spiritual values. The results underline the importance of preserving this traditional knowledge in order to promote honey farming and enhance the value of local knowledge in Africa.
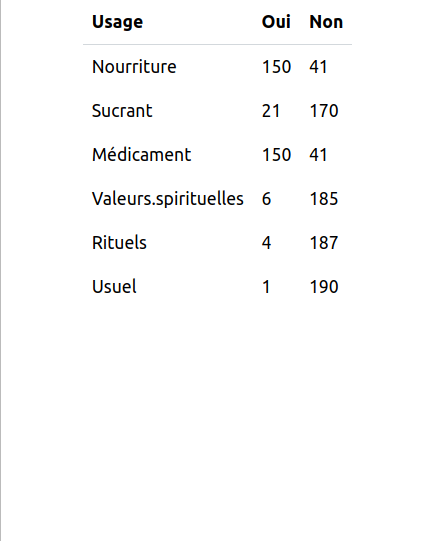
I've also attached an overview of my database of honey uses by species. I also have other data, but not yet in csv format. I have personal information on the people interviewed, such as their age, ethnicity, level of education, where they live, etc.
I know it's possible to find the answer to these questions without using statistics, but it's compulsory to use them at my university. I'm willing to add research questions to incorporate statistics. I'd like to find a way of highlighting the relationship between certain species and certain uses of honey. Or a link between the age, level of education, etc. of the participants and their knowledge. Linking the amount of medical knowledge in a village to the distance to the nearest hospital. I am open to any proposal!
Thank you so much
> dput(mydata)
structure(list(Samples = c("Dactylurina sp. 1", "Dactylurina sp. 2",
"Dactylurina sp. 3", "Dactylurina sp. 4", "Dactylurina sp. 5",
"Dactylurina sp. 6", "Dactylurina sp. 7", "Dactylurina sp. 8",
"Dactylurina sp. 9", "Dactylurina sp. 10", "Dactylurina sp. 11",
"Dactylurina sp. 12", "Dactylurina sp. 13", "Dactylurina sp. 14",
"Dactylurina sp. 15", "Dactylurina sp. 16", "Dactylurina sp. 17",
"Dactylurina sp. 18", "Dactylurina sp. 19", "Dactylurina sp. 20",
"Dactylurina sp. 21", "Dactylurina sp. 22", "Dactylurina sp. 23",
"Dactylurina sp. 24", "Dactylurina sp. 25", "Dactylurina sp. 26",
"Dactylurina sp. 27", "Dactylurina sp. 28", "Dactylurina sp. 29",
"Dactylurina sp. 30", "Dactylurina sp. 31", "Dactylurina sp. 32",
"Plebeina armata 1", "Plebeina armata 2", "Plebeina armata 3",
"Plebeina armata 4", "Plebeina armata 5", "Plebeina armata 6",
"Hypo/liotrigona spp. 1", "Hypo/liotrigona spp. 2", "Hypo/liotrigona spp. 3",
"Hypo/liotrigona spp. 4", "Hypo/liotrigona spp. 5", "Hypo/liotrigona spp. 6",
"Hypo/liotrigona spp. 7", "Hypo/liotrigona spp. 8", "Hypo/liotrigona spp. 9",
"Hypo/liotrigona spp. 10", "Hypo/liotrigona spp. 11", "Hypo/liotrigona spp. 12",
"Hypo/liotrigona spp. 13", "Hypo/liotrigona spp. 14", "Hypo/liotrigona spp. 15",
"Hypo/liotrigona spp. 16", "Hypo/liotrigona spp. 17", "Hypo/liotrigona spp. 18",
"Hypo/liotrigona spp. 19", "Hypo/liotrigona spp. 20", "Hypo/liotrigona spp. 21",
"Hypo/liotrigona spp. 22", "Hypo/liotrigona spp. 23", "Hypo/liotrigona spp. 24",
"Hypo/liotrigona spp. 25", "Hypo/liotrigona spp. 26", "Hypo/liotrigona spp. 27",
"Hypo/liotrigona spp. 28", "Hypo/liotrigona spp. 29", "Hypo/liotrigona spp. 30",
"Hypo/liotrigona spp. 31", "Hypo/liotrigona spp. 32", "Hypo/liotrigona spp. 33",
"Hypo/liotrigona spp. 34", "Hypo/liotrigona spp. 35", "Hypo/liotrigona spp. 36",
"Hypo/liotrigona spp. 37", "Hypo/liotrigona spp. 38", "Hypo/liotrigona spp. 39",
"Hypo/liotrigona spp. 40", "Hypo/liotrigona spp. 41", "Hypo/liotrigona spp. 42",
"Hypo/liotrigona spp. 43", "Hypo/liotrigona spp. 44", "Hypo/liotrigona spp. 45",
"Hypo/liotrigona spp. 46", "Hypo/liotrigona spp. 47", "Hypo/liotrigona spp. 48",
"Hypo/liotrigona spp. 49", "Hypo/liotrigona spp. 50", "Hypo/liotrigona spp. 51",
"Hypo/liotrigona spp. 52", "Hypo/liotrigona spp. 53", "Hypo/liotrigona spp. 54",
"Hypo/liotrigona spp. 55", "Hypo/liotrigona spp. 56", "Hypo/liotrigona spp. 57",
"Hypo/liotrigona spp. 58", "Hypo/liotrigona spp. 59", "Hypo/liotrigona spp. 60",
"Hypo/liotrigona spp. 61", "Meliponula togoensis 1", "Meliponula togoensis 2",
"Meliponula togoensis 3", "Meliponula togoensis 4", "Meliponula togoensis 5",
"Meliponula togoensis 6", "Meliponula togoensis 7", "Meliponula togoensis 8",
"Meliponula togoensis 9", "Meliponula togoensis 10", "Meliponula togoensis 11",
"Meliponula togoensis 12", "Meliponula togoensis 13", "Meliponula togoensis 14",
"Meliponula togoensis 15", "Meliponula togoensis 16", "Meliponula togoensis 17",
"Meliponula togoensis 18", "Meliponula togoensis 19", "Meliponula togoensis 20",
"Meliponula togoensis 21", "Meliponula togoensis 22", "Meliponula togoensis 23",
"Meliponula togoensis 24", "Meliponula togoensis 25", "Meliponula togoensis 26",
"Meliponula togoensis 27", "Meliponula togoensis 28", "Meliponula togoensis 29",
"Meliponula togoensis 30", "Meliponula togoensis 31", "Meliponula togoensis 32",
"Meliponula togoensis 33", "Meliponula togoensis 34", "Meliponula togoensis 35",
"Meliponula togoensis 36", "Meliponula togoensis 37", "Meliponula togoensis 38",
"Meliponula togoensis 39", "Meliponula togoensis 40", "Meliponula togoensis 41",
"Meliponula togoensis 42", "Meliponula togoensis 43", "Meliponula togoensis 44",
"Meliponula togoensis 45", "Meliponula togoensis 46", "Meliponula togoensis 47",
"Meliponula togoensis 48", "Meliponula togoensis 49", "Meliponula togoensis 50",
"Meliponula togoensis 51", "Meliponula bocandei 1", "Meliponula bocandei 2",
"Meliponula bocandei 3", "Meliponula bocandei 4", "Meliponula bocandei 5",
"Meliponula bocandei 6", "Meliponula bocandei 7", "Meliponula bocandei 8",
"Meliponula bocandei 9", "Meliponula bocandei 10", "Meliponula bocandei 11",
"Meliponula bocandei 12", "Meliponula bocandei 13", "Meliponula bocandei 14",
"Meliponula bocandei 15", "Meliponula bocandei 16", "Meliponula bocandei 17",
"Meliponula bocandei 18", "Meliponula bocandei 19", "Meliponula bocandei 20",
"Meliponula bocandei 21", "Meliponula bocandei 22", "Meliponula bocandei 23",
"Meliponula bocandei 24", "Meliponula bocandei 25", "Meliponula bocandei 26",
"Meliponula bocandei 27", "Meliponula bocandei 28", "Meliponula bocandei 29",
"Meliponula bocandei 30", "Meliponula bocandei 31", "Meliponula bocandei 32",
"Meliponula bocandei 33", "Meliponula bocandei 34", "Meliponula bocandei 35",
"Meliponula bocandei 36", "Meliponula bocandei 37", "Meliponula bocandei 38",
"Meliponula bocandei 39", "Meliponula bocandei 40", "Meliponula bocandei 41"
), Species = c("Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina",
"Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina",
"Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina",
"Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina",
"Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina",
"Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina", "Dactylurina",
"Dactylurina", "Dactylurina", "Dactylurina", "Plebeina", "Plebeina",
"Plebeina", "Plebeina", "Plebeina", "Plebeina", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona", "HypoLiotrigona",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_togoensis", "Meliponula_togoensis", "Meliponula_togoensis",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei", "Meliponula_bocandei",
"Meliponula_bocandei", "Meliponula_bocandei"), Nourriture = c(1L,
1L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 1L, 0L, 1L, 1L,
1L, 1L, 1L, 1L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
0L, 0L, 0L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 1L, 0L, 1L, 1L, 0L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 1L,
1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 0L,
0L, 0L, 0L, 1L, 1L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), Sucrant = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 1L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L,
0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L,
0L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 1L, 1L, 1L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 1L,
1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 1L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L), Médicament = c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 1L,
1L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 0L, 0L, 1L, 1L, 0L, 0L,
1L, 1L, 0L, 1L, 1L, 0L, 0L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L, 0L,
1L, 0L, 0L, 1L, 1L, 0L, 1L, 1L, 0L, 1L, 1L, 0L, 1L, 0L, 0L, 0L,
1L, 1L, 0L, 0L, 1L, 1L, 0L, 1L, 1L, 1L, 0L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 1L, 0L, 0L, 1L, 1L, 0L, 1L, 0L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
0L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), Valeurs.spirituelles = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 1L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L), Rituels = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L), Usuel = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L)), class = "data.frame", row.names = c(NA,
-191L))
![]()