Hello, welcome to RStudio Community! First of all, congratulations to go above and beyond and try to learn some R - the initial difficulty curve will be worth it as it will make future work so much easier!
When posting here, it is useful to provide a reproducible example - we can't copy-paste a screenshot of data, so we are limited in how we can help. I'm going to make a fake version of your data on my machine:
library(tidyverse)
dat = crossing(site = c("bl", "bl_c", "c"),
species = c("betula", "salix", "larix")) |>
mutate(amount = rnorm(9, 2000, 500))
# A tibble: 9 x 3
site species amount
<chr> <chr> <dbl>
1 bl betula 1614.
2 bl larix 1686.
3 bl salix 1963.
4 bl_c betula 1687.
5 bl_c larix 1311.
6 bl_c salix 1784.
7 c betula 2251.
8 c larix 2175.
9 c salix 2330.
So with the data like this, if I want to know the percentage of each species in each forest, I'd group by the the factor where, within each value, the "percentage" column should add up to 100%. Then, I mutate a new column which is equal to the value column divided by the sum of the value column. As the data frame is grouped, the sum of the value column is just the sum of that given group.
dat |>
group_by(site) |>
mutate(perc = amount / sum(amount))
# A tibble: 9 x 4
# Groups: site [3]
site species amount perc
<chr> <chr> <dbl> <dbl>
1 bl betula 1427. 0.296
2 bl larix 1696. 0.352
3 bl salix 1700. 0.352
4 bl_c betula 2104. 0.328
5 bl_c larix 2368. 0.369
6 bl_c salix 1942. 0.303
7 c betula 2965. 0.392
8 c larix 2236. 0.296
9 c salix 2364. 0.312
The way to test this could now be to sum up the percentage column - each should be 100% - which it is!
dat |>
group_by(site) |>
mutate(perc = amount / sum(amount)) |>
summarise(sum(perc))
# A tibble: 3 x 2
site `sum(perc)`
<chr> <dbl>
1 bl 1
2 bl_c 1
3 c 1
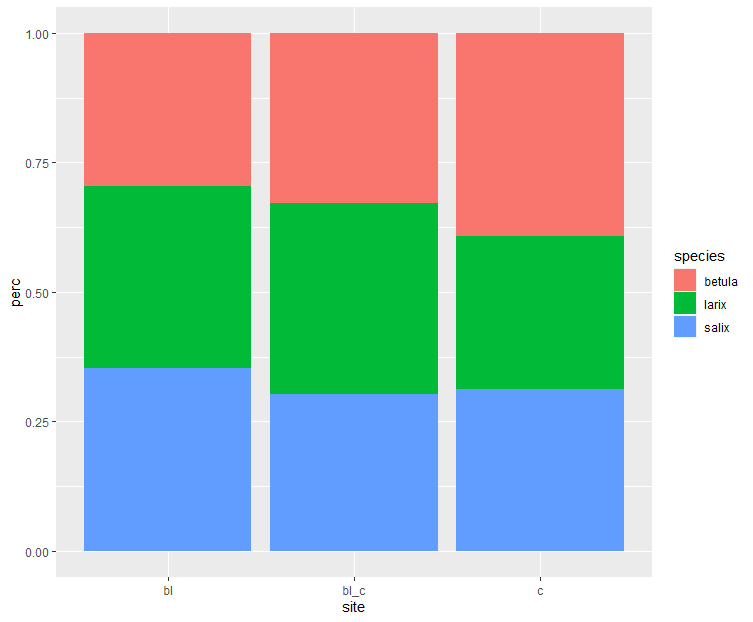
Or we could visualise it...
dat |>
group_by(site) |>
mutate(perc = amount / sum(amount)) |>
ggplot(aes(x = site, y = perc, fill = species)) +
geom_col()
NB: If you want to get rid of NA values, you could drop them by doing something like:
dat |>
drop_na(species)