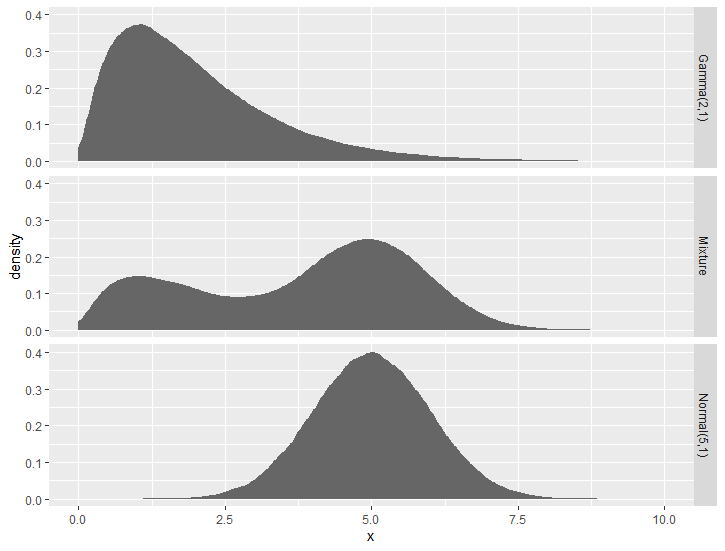
One simple way is to reform the data, because normal geom_* functions won't able to recognize the distribution syntax.
df1 <- df %>% rowwise %>% mutate(x = list(generate(dist, 100000))) %>% unnest_longer(x)
Then it is easy to draw a distribution using stat = "density" in geom_*() functions.
ggplot(df1) +
geom_area(aes(x = x, group = name), stat = "density", fill = "grey40")+
facet_grid(rows = vars(name)) +
xlim(0,10)
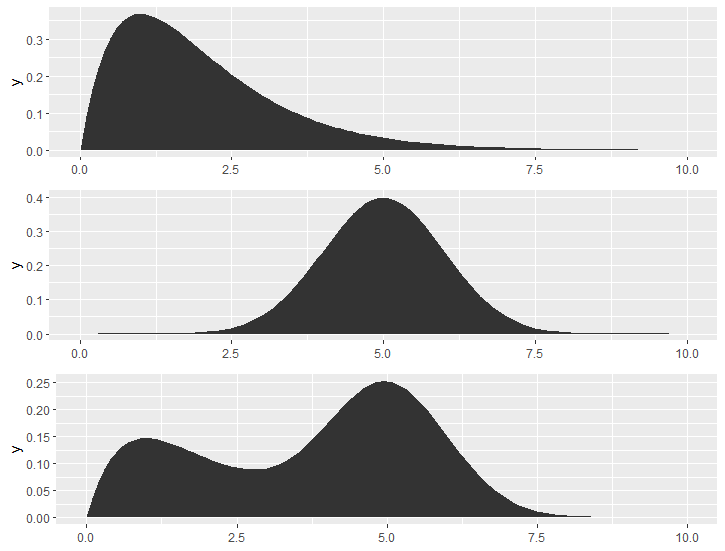
If you want perfect smooth line for these distribution curves, you may consider directly draw the density function using stat_function(). but I yet don't know how to vertically parallelly draw the 3 _function layers with only using ggplot2 functions, may be require modifying ggproto(), or looking for help from plot_grid(), but that's too complicated.
library(cowplot)
gama_dis <- ggplot() +
stat_function(fun = \(x) dgamma(x, shape = 2, rate = 1),geom = "polygon") +
xlim(0, 10)
normal_dis <- ggplot() +
stat_function(fun = \(x) dnorm(x, mean = 5, sd = 1), geom = "polygon") +
xlim(0,10)
mix_dis <- ggplot() +
stat_function(fun = \(x) 0.4 * dgamma(x, shape = 2, rate = 1) + 0.6 * dnorm(x, mean = 5, sd = 1),
geom = "polygon") +
xlim(0,10)
plot_grid(gama_dis, normal_dis, mix_dis, ncol = 1)