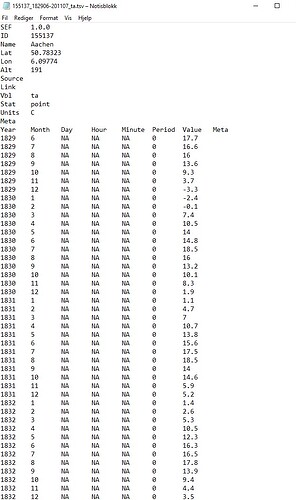
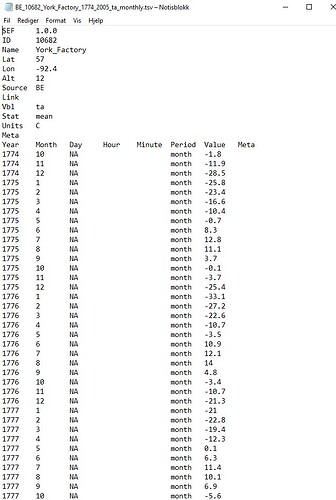
Actually no because we lose the ID in the file. I think I have a simple hack of the write_sef function that gives us what we want.
I simply changed the line
filename <- paste(sou, cod, dates, variable, sep = "_")
to
filename <- paste(sou, cod, nam, dates, variable, sep = "_")
and saved the function under the name play and it seems to work,
Paste the function below into R and
**New Function play. **
play <- function (Data, outpath, variable, cod, nam = "", lat = "", lon = "",
alt = "", sou = "", link = "", units, stat, metaHead = "",
meta = "", period = "", time_offset = 0, note = "", keep_na = FALSE,
outfile = NA)
{
for (i in 1:ncol(Data)) Data[, i] <- as.character(Data[,
i])
header <- array(dim = c(12, 2), data = "")
header[1, ] <- c("SEF", "1.0.0")
header[2, ] <- c("ID", trimws(as.character(cod)))
header[3, ] <- c("Name", trimws(as.character(nam)))
header[4, ] <- c("Lat", trimws(as.character(lat)))
header[5, ] <- c("Lon", trimws(as.character(lon)))
header[6, ] <- c("Alt", trimws(as.character(alt)))
header[7, ] <- c("Source", trimws(as.character(sou)))
header[8, ] <- c("Link", trimws(as.character(link)))
header[9, ] <- c("Vbl", trimws(as.character(variable)))
header[10, ] <- c("Stat", trimws(as.character(stat)))
header[11, ] <- c("Units", trimws(as.character(units)))
header[12, ] <- c("Meta", trimws(as.character(metaHead)))
if (stat == "point" & !all(as.character(period) == "0")) {
period <- "0"
warning("Period forced to 0 because of 'stat'")
}
if (!all(time_offset == 0) & !all(is.na(as.integer(Data[,
4]) + as.integer(Data[, 5])))) {
times <- ISOdate(Data[, 1], Data[, 2], Data[, 3], Data[,
4], Data[, 5])
times <- times - time_offset * 3600
Data[which(!is.na(times)), 1] <- as.integer(substr(times[which(!is.na(times))],
1, 4))
Data[which(!is.na(times)), 2] <- as.integer(substr(times[which(!is.na(times))],
6, 7))
Data[which(!is.na(times)), 3] <- as.integer(substr(times[which(!is.na(times))],
9, 10))
Data[which(!is.na(times)), 4] <- as.integer(substr(times[which(!is.na(times))],
12, 13))
Data[which(!is.na(times)), 5] <- as.integer(substr(times[which(!is.na(times))],
15, 16))
}
DataNew <- data.frame(Year = Data[, 1], Month = Data[, 2],
Day = Data[, 3], Hour = Data[, 4], Minute = Data[, 5],
Period = as.character(period), Value = Data[, 6], Meta = as.character(meta),
stringsAsFactors = FALSE)
if (!keep_na)
DataNew <- DataNew[which(!is.na(DataNew$Value)), ]
if (substr(outpath, nchar(outpath), nchar(outpath)) != "/") {
outpath <- paste0(outpath, "/")
}
if (is.na(outfile)) {
j <- 3
if (is.na(as.integer(DataNew[1, 3])))
j <- 2
if (is.na(as.integer(DataNew[1, 2])))
j <- 1
datemin <- paste(formatC(unlist(as.integer(DataNew[1,
1:j])), width = 2, flag = 0), collapse = "")
datemax <- paste(formatC(unlist(as.integer(DataNew[dim(DataNew)[1],
1:j])), width = 2, flag = 0), collapse = "")
dates <- paste(datemin, datemax, sep = "-")
filename <- paste(sou, cod, nam, dates, variable, sep = "_")
if (sou %in% c(NA, ""))
filename <- sub("_", "", filename)
if (note != "") {
note <- paste0("_", gsub(" ", "_", note))
}
filename <- gsub(" ", "", filename)
filename <- paste0(outpath, filename, note, ".tsv")
}
else {
filename <- paste0(outpath, outfile)
if (substr(filename, nchar(filename) - 3, nchar(filename)) !=
".tsv") {
filename <- paste0(filename, ".tsv")
}
}
write.table(header, file = filename, quote = FALSE, row.names = FALSE,
col.names = FALSE, sep = "\t", dec = ".", fileEncoding = "UTF-8")
write.table(t(names(DataNew)), file = filename, quote = FALSE,
row.names = FALSE, col.names = FALSE, sep = "\t", fileEncoding = "UTF-8",
append = TRUE)
write.table(DataNew, file = filename, quote = FALSE, row.names = FALSE,
col.names = FALSE, sep = "\t", dec = ".", fileEncoding = "UTF-8",
append = TRUE)
message(paste("Data written to file", filename))
}
and run this slightly revised code.
setwd("/home/john/RJunk/elinlun")
library(tidyverse)
library(dataresqc)
inventory <- read.csv("Inventory_Berkeley_Earth.csv", sep = "\t", header = FALSE)
colnames(inventory) <- tolower(c("Other_ID", "City", "Modern_Country", "Lat.degN", "Lon.degE",
"Station_Elevation.m", "Start_Year", "End_Year" ))
allfiles = list.files("/home/john/RJunk/elinlun/allfiles", full.names=TRUE)
new_inventory <- arrange(inventory, city)
for(i in 1:nrow(new_inventory)){
print(i)
dat1 <- read.csv(allfiles[i], sep = "\t", header = FALSE)
names(dat1) <- c("year", "month", "value")
nought <- data.frame(day = "", hour = "" , minute = "")
dat1 <- cbind(dat1, nought)
dat1 <- dat1[, c("year", "month", "day", "hour", "minute", "value")]
outpath <- "/home/john/RJunk/elinlun/output"
variable = "ta"
lat <- new_inventory[i,][4]
lon <- new_inventory[i,][5]
alt <- new_inventory[i,][6]
cod <- new_inventory[i,][1]
nam <- new_inventory[i,][2]
stat = "mean"
units <- "C"
sou <- "Berkeley_Earth"
note = "monthly"
play(dat1, outpath = outpath, variable = variable, cod = cod, nam = nam,
lat = lat, lon = lon, alt = alt, sou = sou, stat = stat, units = "C", note = note)
}
Note that the only difference is write_sef has changed to play.
![]() (Nobody said R was rationale by programming standards.
(Nobody said R was rationale by programming standards.