See the FAQ: How to do a minimal reproducible example reprex for beginners for a more specific answer.
See if you can run the examples from help(compana)
suppressPackageStartupMessages({
library(adehabitatHS)
})
## The examples presented here
## are the same as those presented in
## the paper of Aebischer et al. (1993)
#############################
## Pheasant dataset: first
## example in Aebischer et al.
data(pheasant)
## Second order habitat selection
## Selection of home range within the
## study area (example of parametric test)
pheana2 <- compana(pheasant$mcp, pheasant$studyarea,
test = "parametric")
pheana2
#> ************ Compositional analysis of habitat use ***************
#>
#> The analysis was carried out with 13 animals and 5 habitat types
#> 1. Test of the habitat selection:
#> parametric test
#> Lambda df P
#> 8.491656e-02 4.000000e+00 1.860604e-06
#>
#> 2. Ranking of habitats (profile):
#>
#> habitat Scrub Broadleaf Grassland Coniferous Crop
#> Scrub ------------------
#> Broadleaf ------------------
#> Grassland -----------------------
#> Coniferous -----------------------
#> Crop ------
## The ranking matrix:
print(pheana2$rm, quote = FALSE)
#> Scrub Broadleaf Coniferous Grassland Crop
#> Scrub 0 + +++ +++ +++
#> Broadleaf - 0 +++ +++ +++
#> Coniferous --- --- 0 - +++
#> Grassland --- --- + 0 +++
#> Crop --- --- --- --- 0
## Third order habitat selection
## (relocation within home range)
## We remove the first pheasant of the analysis
## (as in the paper of Aebischer et al.)
## before the analysis
pheana3 <- compana(pheasant$locs[-1,], pheasant$mcp[-1,c(1,2,4)])
pheana3
#> ************ Compositional analysis of habitat use ***************
#>
#> The analysis was carried out with 12 animals and 3 habitat types
#> 1. Test of the habitat selection:
#> randomisation test
#> Lambda P
#> 0.3655733 0.0060000
#>
#> 2. Ranking of habitats (profile):
#>
#> habitat Broadleaf Scrub Grassland
#> Broadleaf ------------------
#> Scrub ------------------
#> Grassland -----------
## The ranking matrix:
print(pheana3$rm, quote = FALSE)
#> Scrub Broadleaf Grassland
#> Scrub 0 - +++
#> Broadleaf + 0 +++
#> Grassland --- --- 0
#############################
## Squirrel data set: second
## example in Aebischer et al.
data(squirrel)
## Second order habitat selection
## Selection of home range within the
## study area
squiana2 <- compana(squirrel$mcp, squirrel$studyarea)
squiana2
#> ************ Compositional analysis of habitat use ***************
#>
#> The analysis was carried out with 17 animals and 5 habitat types
#> 1. Test of the habitat selection:
#> randomisation test
#> Lambda P
#> 0.06808452 0.00200000
#>
#> 2. Ranking of habitats (profile):
#>
#> habitat Larch Mature Young Open Thuja
#> Larch ----------------------
#> Mature ----------------------
#> Young ----------------------------
#> Open -------------
#> Thuja -------
## The ranking matrix:
print(squiana2$rm, quote = FALSE)
#> Young Thuja Larch Mature Open
#> Young 0 +++ - - +
#> Thuja --- 0 --- --- ---
#> Larch + +++ 0 + +++
#> Mature + +++ - 0 +++
#> Open - +++ --- --- 0
## However, note that here, the hypothesis of identical use
## on which this analysis relies is likely to be false.
## Indeed, an eisera indicates:
us <- round(30 * squirrel$locs / 100)
av <- squirrel$studyarea
ii <- eisera(us, av, scannf = FALSE)
scatter(ii, grid = FALSE, clab = 0.7)
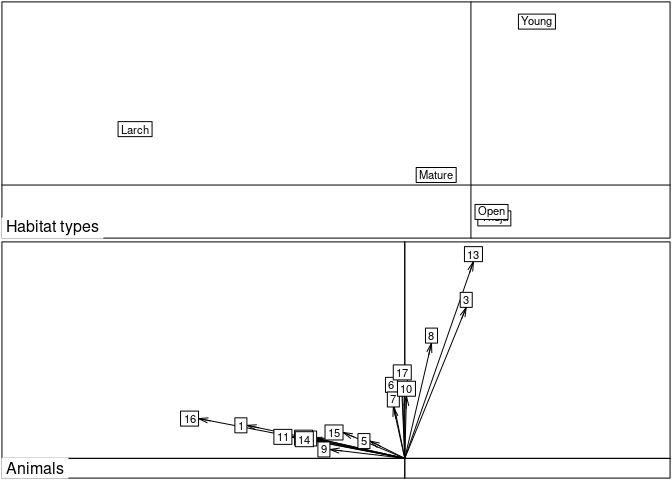
## There are clearly two groups of animals. In such cases,
## compositional analysis is to be avoided in this case.