Hi @MLaure19, I've reconstructed the city names from the previous data you posted, and have something you could now try, but it depends on restructuring your data into long form, which is often convenient for visualizations. (Also this is a reprex, so anyone can copy, paste and run the code, since it has the data, the packages used, and the code.)
load data
### data
my_table <-
structure(list(NOM_IRIS = structure(c(18L, 127L, 23L, 89L, 28L,
103L, 87L, 41L, 26L, 175L, 42L, 43L, 40L, 171L, 102L, 101L, 159L,
63L, 99L, 98L), .Label = c("Abbaye", "Abry", "Aigle", "AlliŽs-Clos d'Or",
"Alpins", "Ambroise Croizat", "Arc-En-Ciel", "Aristide Briand",
"Arlequin", "Ayguinards", "Bajati\217re Est", "Bajati\217re Ouest",
"Baladins", "Barnave-Saint-Robert", "Bastille", "Beauvert", "Berriat-Amp\217re",
"Bresson", "BŽali\217res", "Capuche", "Champaviotte", "Championnet",
"Claix Centre Bourg", "Clemenceau", "Clinique Mutualiste", "Comboire",
"Constantine", "Corenc", "Cours Berriat", "Croix Rouge", "CrŽqui-Victor Hugo",
"Curie-Rolland", "Diables Bleus", "Diderot", "Domaine Universitaire",
"Drac-Ampere", "Drac-Vercors", "Eaux Claires-Champs Elysees",
"Eaux Claires-PainlevŽ", "Echirolles Centre", "Echirolles Ouest-Nord",
"Echirolles Ouest-Sud", "Echirolles Za du Centre", "Engenieres-Rolandiere",
"Esplanade", "Europole", "Eybens Nord", "Eybens Sud Est", "Eybens Sud Ouest",
"FerriŽ-Stalingrad", "Fiancey-Brieux", "Foch Est", "Foch Ouest",
"Fontaine Mairie", "Fontaine Zone Industrielle", "Fontanil-Cornillon",
"Frange Verte-Commanderie", "Gabriel PŽri", "Gare", "Gi\217res Nord",
"Gi\217res Sud", "Glairons", "Grand-Place", "Grand Place Alpexpo",
"Grand Pre-Buclos-Eyminees", "Grenette", "Gringalet", "Gustave Rivet",
"GŽnissieu", "Haut de la Commune", "Haut Meylan", "Hebert-Mutualite",
"Helbronner-Geants", "Hoche", "Houille Blanche", "Ile Verte-MarŽchal Randon",
"Ile Verte-Saint Roch", "Jaur\217s-Vallier", "Jean Jaures", "Jean MacŽ",
"Jeanne d'Arc", "Jouhaux", "La Bruyere", "La Falaise", "La Fauconni\217re",
"La Monta-Visancourt", "Le Bourg-Les Chenevi\217res-Chantavot-Zone Indu",
"Le Bourg-Les C\231tes", "Le Coteau", "Le Gua", "Le Pont-de-Claix Zone d'ActivitŽs des ëles",
"Les Alloves", "Les Alpes", "Les Buissonnees", "Les ëles de Mars-Les Olympiades",
"Les ëles Nord", "Les ëles Sud", "Les Essarts-Nord", "Les Essarts-Sud",
"Les Floralies", "Les Granges-Nord", "Les Granges-Sud", "Les Mortes l'Oratoire Les Contamines",
"Les Trembles", "Les Vouillands", "Lustucru", "Mairie-Langevin",
"Malherbe", "Maquis du Gresivaudan-Nord", "Maquis du Gresivaudan-Sud",
"Maupertuis-St Mury-Charlaix", "Meylan Zone d'ActivitŽs 1", "Meylan Zone d'ActivitŽs 2",
"Meylan Zone d'ActivitŽs 3", "Mistral", "Moutonnees", "Murianette",
"Mžrier", "Neyrpic", "Notre Dame", "Noyarey", "Pacalaire", "Paul-Eluard",
"Paul Bert", "Paul Cocat", "Peretto", "Plaine du Pont Rouge",
"Plaine Fleurie", "Plateforme Chimique", "Poisat", "Polygone",
"Pont-de-Claix Bourg", "Portail Rouge", "Poterne", "Poya-Plans",
"Predieu", "PrŽfecture", "Renaudie", "Reviree", "Reynies", "Rochepleine",
"Rondeau-Liberation", "Saint-Andre", "Saint-Bruno", "Saint-ƒgr\217ve Zone Industrielle",
"Saint-Laurent-Lavalette", "Saint-Martin-d'H\217res La Plaine",
"Saint-Martin-d'H\217res Le Village", "Saint-Martin-le-Vinoux Nord Est",
"Saint-Martin-le-Vinoux Sud Ouest et ZI", "Saint-Paul-de-Varces",
"Sassenage La Plaine", "Semard", "Seyssinet-Pariset Le Village",
"Seyssins La Plaine", "Seyssins Le Centre", "Seyssins Le Village",
"Sidi-Brahim", "Surieux", "Taillees", "Teisseire", "Trois Cours",
"Vallier", "Valmy", "Varces Centre", "Varces PŽriphŽrie", "Veurey-Voroize",
"Vif Centre Ville", "Vif PŽriphŽrie", "Vigny-Musset", "Village 2",
"Village Olympique Nord", "Village Olympique Sud", "Villancourt",
"Viscose-Luire", "Waldeck-Rousseau", "Zone SpŽcifique l'ële d'Amour"
), class = "factor"), HISTO_18.3 = c(208L, 233L, 39L, 2L, 802L,
83L, 385L, 0L, 53L, 0L, 2L, 0L, 271L, 29L, 0L, 0L, 0L, 0L, 0L,
0L), HISTO_18.4 = c(182L, 214L, 14L, 105L, 689L, 88L, 258L, 0L,
54L, 2L, 21L, 0L, 230L, 28L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_18.5 = c(173L,
184L, 14L, 107L, 570L, 57L, 336L, 0L, 39L, 10L, 67L, 0L, 253L,
32L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_18.6 = c(126L, 178L, 17L,
38L, 450L, 67L, 268L, 0L, 35L, 42L, 77L, 0L, 231L, 24L, 0L, 0L,
0L, 0L, 0L, 0L), HISTO_18.7 = c(115L, 123L, 8L, 33L, 465L, 34L,
284L, 0L, 44L, 84L, 79L, 0L, 184L, 18L, 0L, 0L, 0L, 0L, 0L, 0L
), HISTO_18.8 = c(104L, 182L, 10L, 0L, 435L, 31L, 173L, 0L, 37L,
135L, 70L, 0L, 166L, 24L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_18.9 = c(104L,
158L, 14L, 0L, 366L, 33L, 240L, 0L, 88L, 146L, 48L, 0L, 155L,
25L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_19 = c(84L, 147L, 16L, 0L,
328L, 20L, 192L, 0L, 60L, 136L, 50L, 0L, 135L, 20L, 0L, 0L, 0L,
0L, 0L, 0L), HISTO_19.1 = c(100L, 139L, 4L, 0L, 314L, 16L, 110L,
0L, 49L, 111L, 93L, 0L, 172L, 14L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_19.2 = c(83L,
112L, 18L, 0L, 286L, 20L, 138L, 0L, 34L, 110L, 97L, 0L, 215L,
11L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_19.3 = c(75L, 136L, 11L,
0L, 266L, 23L, 141L, 0L, 51L, 117L, 72L, 0L, 240L, 19L, 0L, 0L,
0L, 0L, 0L, 0L), HISTO_19.4 = c(65L, 142L, 10L, 0L, 230L, 18L,
101L, 0L, 50L, 107L, 70L, 0L, 211L, 19L, 0L, 0L, 0L, 0L, 0L,
0L), HISTO_19.5 = c(64L, 100L, 8L, 0L, 219L, 19L, 97L, 0L, 39L,
107L, 62L, 41L, 211L, 13L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_19.6 = c(69L,
118L, 15L, 36L, 218L, 15L, 93L, 0L, 44L, 76L, 49L, 104L, 189L,
6L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_19.7 = c(41L, 90L, 4L, 19L,
149L, 36L, 87L, 0L, 37L, 94L, 55L, 79L, 159L, 10L, 0L, 0L, 0L,
0L, 0L, 0L), HISTO_19.8 = c(64L, 87L, 9L, 40L, 153L, 16L, 99L,
0L, 42L, 94L, 50L, 73L, 161L, 7L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_19.9 = c(30L,
119L, 9L, 0L, 126L, 55L, 107L, 0L, 35L, 80L, 49L, 75L, 182L,
10L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_20 = c(28L, 88L, 10L, 16L,
121L, 10L, 54L, 0L, 35L, 78L, 50L, 69L, 167L, 8L, 0L, 0L, 0L,
0L, 0L, 0L), HISTO_20.1 = c(42L, 53L, 3L, 0L, 113L, 28L, 71L,
0L, 31L, 70L, 42L, 72L, 214L, 8L, 0L, 0L, 0L, 0L, 0L, 0L), HISTO_20.2 = c(23L,
66L, 6L, 2L, 97L, 20L, 57L, 0L, 29L, 77L, 20L, 69L, 240L, 7L,
0L, 0L, 0L, 0L, 0L, 0L), HISTO_20.3 = c(24L, 65L, 4L, 10L, 91L,
7L, 36L, 0L, 27L, 76L, 39L, 76L, 246L, 7L, 0L, 0L, 0L, 0L, 0L,
0L)), row.names = c(NA, -20L), class = c("tbl_df", "tbl", "data.frame"
))
### end of 'structure()' command
### load packages needed to run code
library(tidyverse)
### code
# inspect table
my_table %>% print(n = 6)
#> # A tibble: 20 x 22
#> NOM_IRIS HISTO_18.3 HISTO_18.4 HISTO_18.5 HISTO_18.6 HISTO_18.7
#> <fct> <int> <int> <int> <int> <int>
#> 1 Bresson 208 182 173 126 115
#> 2 Plaine … 233 214 184 178 123
#> 3 Claix C… 39 14 14 17 8
#> 4 Le Cote… 2 105 107 38 33
#> 5 Corenc 802 689 570 450 465
#> 6 Les Mor… 83 88 57 67 34
#> # … with 14 more rows, and 16 more variables: HISTO_18.8 <int>,
#> # HISTO_18.9 <int>, HISTO_19 <int>, HISTO_19.1 <int>, HISTO_19.2 <int>,
#> # HISTO_19.3 <int>, HISTO_19.4 <int>, HISTO_19.5 <int>,
#> # HISTO_19.6 <int>, HISTO_19.7 <int>, HISTO_19.8 <int>,
#> # HISTO_19.9 <int>, HISTO_20 <int>, HISTO_20.1 <int>, HISTO_20.2 <int>,
#> # HISTO_20.3 <int>
# 'long' version of table
my_table %>%
mutate(NOM_IRIS = NOM_IRIS)%>%
pivot_longer(-NOM_IRIS) %>%
# extract numbers from column names
# (I choose to name the column 'pollution' for 'level of pollution')
mutate(pollution = parse_number(name)) %>% print(n = 6)
#> # A tibble: 420 x 4
#> NOM_IRIS name value pollution
#> <fct> <chr> <int> <dbl>
#> 1 Bresson HISTO_18.3 208 18.3
#> 2 Bresson HISTO_18.4 182 18.4
#> 3 Bresson HISTO_18.5 173 18.5
#> 4 Bresson HISTO_18.6 126 18.6
#> 5 Bresson HISTO_18.7 115 18.7
#> 6 Bresson HISTO_18.8 104 18.8
#> # … with 414 more rows
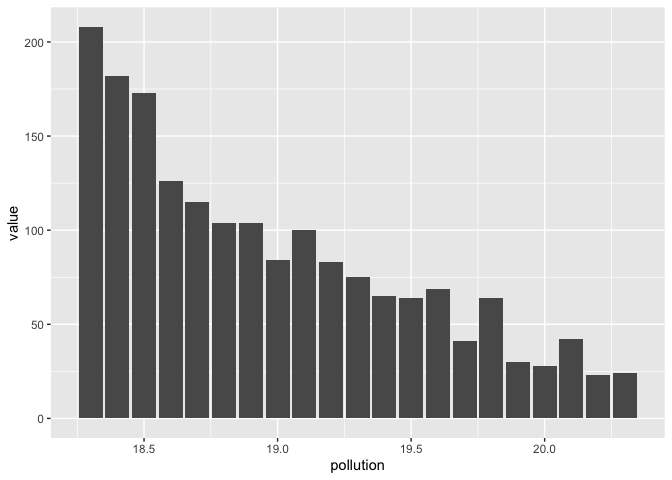
# extract Bresson data and plot histogram
my_table %>%
mutate(NOM_IRIS = NOM_IRIS)%>%
pivot_longer(-NOM_IRIS) %>%
mutate(pollution = parse_number(name)) %>%
# extract Bresson data
filter(NOM_IRIS == 'Bresson') %>%
# plot histogram
ggplot(aes(pollution, value)) +
geom_col()
Created on 2020-03-31 by the reprex package (v0.3.0)
Edit: I mixed two versions of the code, but have made it consistent now.